7AZS
 
 | | 70S thermus thermophilus ribosome with bound antibiotic lead SEQ-569 | | Descriptor: | (2R,3S,4R,5R,7S,9S,10S,11R,12S,13R)-12-(((2R,4R,5S,6S)-4,5-dihydroxy-4,6-dimethyltetrahydro-2H-pyran-2-yl)oxy)-2-((S)-1-(((2R,3R,4R,5R,6R)-5-hydroxy-3,4-dimethoxy-6-methyltetrahydro-2H-pyran-2-yl)oxy)propan-2-yl)-10-(((2S,3R,6R,E)-3-hydroxy-4-(methoxyimino)-6-methyltetrahydro-2H-pyran-2-yl)oxy)-3,5,7,9,11,13-hexamethyl-7-(((2-(2-methyl-5-nitro-1H-imidazol-1-yl)ethyl)carbamoyl)oxy)-6,14-dioxooxacyclotetradecan-4-yl 3-methylbutanoate, 16S rRNA, 23S rRNA, ... | | Authors: | Jenner, L.B, Yusupov, M, Yusupova, G, Rak, A. | | Deposit date: | 2020-11-17 | | Release date: | 2022-06-08 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of natural-product-derived sequanamycins as potent oral anti-tuberculosis agents.
Cell, 186, 2023
|
|
4UUQ
 
 | | Crystal structure of human mono-glyceride lipase in complex with SAR127303 | | Descriptor: | 4-({[(4-chlorophenyl)sulfonyl]amino}methyl)piperidine-1-carboxylic acid, MONOGLYCERIDE LIPASE | | Authors: | Griebel, G, Pichat, P, Beeske, S, Leroy, T, Redon, N, Francon, D, Bert, L, Even, L, Lopez-Grancha, M, Tolstykh, T, Sun, F, Yu, Q, Brittain, S, Arlt, H, He, T, Zhang, B, Wiederschain, D, Bertrand, T, Houtman, J, Rak, A, Vallee, F, Michot, N, Auge, F, Menet, V, Bergis, O.E, George, P, Avenet, P, Mikol, V, Didier, M, Escoubet, J. | | Deposit date: | 2014-07-30 | | Release date: | 2015-01-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Selective Blockade of the Hydrolysis of the Endocannabinoid 2-Arachidonoylglycerol Impairs Learning and Memory Performance While Producing Antinociceptive Activity in Rodents.
Sci.Rep., 5, 2015
|
|
5FHX
 
 | | CRYSTAL STRUCTURE OF CODV IN COMPLEX WITH IL4 AT 2.55 Ang. RESOLUTION. | | Descriptor: | Antibody fragment light chain, Interleukin-4, PHOSPHATE ION, ... | | Authors: | Vallee, F, Dupuy, A, Rak, A. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-30 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | CODV-Ig, a universal bispecific tetravalent and multifunctional immunoglobulin format for medical applications.
Mabs, 8, 2016
|
|
1LTX
 
 | | Structure of Rab Escort Protein-1 in complex with Rab geranylgeranyl transferase and isoprenoid | | Descriptor: | AAAA, CHLORIDE ION, FARNESYL, ... | | Authors: | Pylypenko, O, Rak, A, Reents, R, Niculae, A, Thoma, N.H, Waldmann, H, Schlichting, I, Goody, R.S, Alexandrov, K. | | Deposit date: | 2002-05-21 | | Release date: | 2003-05-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Rab Escort Protein-1 in Complex with Rab Geranylgeranyltransferase
Mol.Cell, 11, 2003
|
|
8TYO
 
 | |
8TYL
 
 | |
6SBO
 
 | | Estrogen receptor mutant L536S | | Descriptor: | 6-(2,4-dichlorophenyl)-5-[4-[(3~{S})-1-(3-fluoranylpropyl)pyrrolidin-3-yl]oxyphenyl]-8,9-dihydro-7~{H}-benzo[7]annulene-2-carboxylic acid, Estrogen receptor | | Authors: | Vallee, F, Steier, V, Rak, A. | | Deposit date: | 2019-07-22 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Discovery of 6-(2,4-Dichlorophenyl)-5-[4-[(3S)-1-(3-fluoropropyl)pyrrolidin-3-yl]oxyphenyl]-8,9-dihydro-7H-benzo[7]annulene-2-carboxylic acid (SAR439859), a Potent and Selective Estrogen Receptor Degrader (SERD) for the Treatment of Estrogen-Receptor-Positive Breast Cancer.
J.Med.Chem., 63, 2020
|
|
5BX0
 
 | |
3DAD
 
 | | Crystal structure of the N-terminal regulatory domains of the formin FHOD1 | | Descriptor: | FH1/FH2 domain-containing protein 1 | | Authors: | Schulte, A, Stolp, B, Schonichen, A, Pylypenko, O, Rak, A, Fackler, O.T, Geyer, M. | | Deposit date: | 2008-05-29 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Human Formin FHOD1 Contains a Bipartite Structure of FH3 and GTPase-Binding Domains Required for Activation.
Structure, 16, 2008
|
|
5HCG
 
 | | CRYSTAL STRUCTURE OF FREE-CODV. | | Descriptor: | CODV heavy-chain, CODV light chain | | Authors: | Vallee, F, Dupuy, A, Rak, A. | | Deposit date: | 2016-01-04 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | CODV-Ig, a universal bispecific tetravalent and multifunctional immunoglobulin format for medical applications.
Mabs, 8, 2016
|
|
2BCG
 
 | | Structure of doubly prenylated Ypt1:GDI complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GERAN-8-YL GERAN, GTP-binding protein YPT1, ... | | Authors: | Pylypenko, O, Rak, A, Alexandrov, K. | | Deposit date: | 2005-10-19 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure of doubly prenylated Ypt1:GDI complex and the mechanism of GDI-mediated Rab recycling
Embo J., 25, 2006
|
|
2FU5
 
 | | structure of Rab8 in complex with MSS4 | | Descriptor: | BETA-MERCAPTOETHANOL, Guanine nucleotide exchange factor MSS4, Ras-related protein Rab-8A, ... | | Authors: | Itzen, A, Pylypenko, O, Goody, R.S, Rak, A. | | Deposit date: | 2006-01-26 | | Release date: | 2006-04-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Nucleotide exchange via local protein unfolding-structure of Rab8 in complex with MSS4
Embo J., 25, 2006
|
|
2GIL
 
 | | Structure of the extremely slow GTPase Rab6A in the GTP bound form at 1.8 resolution | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-6A | | Authors: | Bergbrede, T, Pylypenko, O, Rak, A, Alexandrov, K. | | Deposit date: | 2006-03-29 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of the extremely slow GTPase Rab6A in the GTP bound form at 1.8 resolution
J.STRUCT.BIOL., 152, 2005
|
|
2RAI
 
 | | The PX-BAR membrane remodeling unit of Sorting Nexin 9 | | Descriptor: | Sorting nexin-9 | | Authors: | Pylypenko, O, Lundmark, R, Rasmuson, E, Carlsson, S.R, Rak, A. | | Deposit date: | 2007-09-16 | | Release date: | 2007-12-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The PX-BAR membrane-remodeling unit of sorting nexin 9
Embo J., 26, 2007
|
|
2RAJ
 
 | | SO4 bound PX-BAR membrane remodeling unit of Sorting Nexin 9 | | Descriptor: | SULFATE ION, Sorting nexin-9 | | Authors: | Pylypenko, O, Lundmark, R, Rasmuson, E, Carlsson, S.R, Rak, A. | | Deposit date: | 2007-09-16 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The PX-BAR membrane-remodeling unit of sorting nexin 9
Embo J., 26, 2007
|
|
1N88
 
 | | NMR structure of the ribosomal protein L23 from Thermus thermophilus. | | Descriptor: | Ribosomal protein L23 | | Authors: | Ohman, A, Rak, A, Dontsova, M, Garber, M.B, Hard, T. | | Deposit date: | 2002-11-20 | | Release date: | 2003-06-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the ribosomal protein L23 from Thermus thermophilus.
J.Biomol.NMR, 26, 2003
|
|
2RAK
 
 | | PI(3)P bound PX-BAR membrane remodeling unit of Sorting Nexin 9 | | Descriptor: | 2-(BUTANOYLOXY)-1-{[(HYDROXY{[2,3,4,6-TETRAHYDROXY-5-(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL)OXY]METHYL}ETHYL BUTANOATE, Sorting nexin-9 | | Authors: | Pylypenko, O, Lundmark, R, Rasmuson, E, Carlsson, S.R, Rak, A. | | Deposit date: | 2007-09-16 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The PX-BAR membrane-remodeling unit of sorting nexin 9
Embo J., 26, 2007
|
|
3JWE
 
 | | Crystal structure of human mono-glyceride lipase in complex with SAR629 | | Descriptor: | 1-[bis(4-fluorophenyl)methyl]-4-(1H-1,2,4-triazol-1-ylcarbonyl)piperazine, MGLL protein | | Authors: | Bertrand, T, Auge, F, Houtmann, J, Rak, A, Vallee, F, Mikol, V, Berne, P.F, Michot, N, Cheuret, D, Hoornaert, C, Mathieu, M. | | Deposit date: | 2009-09-18 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for human monoglyceride lipase inhibition.
J.Mol.Biol., 396, 2010
|
|
3JW8
 
 | | Crystal structure of human mono-glyceride lipase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, MGLL protein | | Authors: | Bertrand, T, Auge, F, Houtmann, J, Rak, A, Vallee, F, Mikol, V, Berne, P.F, Michot, N, Cheuret, D, Hoornaert, C, Mathieu, M. | | Deposit date: | 2009-09-18 | | Release date: | 2010-01-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for human monoglyceride lipase inhibition.
J.Mol.Biol., 396, 2010
|
|
4AJW
 
 | | Discovery and Optimization of New Benzimidazole- and Benzoxazole-Pyrimidone Selective PI3KBeta Inhibitors for the Treatment of Phosphatase and TENsin homologue (PTEN)-Deficient Cancers | | Descriptor: | 2-[(1-methyl-1H-benzimidazol-2-yl)methyl]-6-morpholin-4-ylpyrimidin-4(3H)-one, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Certal, V, Halley, F, Virone-Oddos, A, Delorme, C, Karlsson, A, Rak, A, Thompson, F, Filoche-Romme, B, El-Ahmad, Y, Carry, J.C, Abecassis, P.Y, Lejeune, P, Bonnevaux, H, Nicolas, J.P, Bertrand, T, Marquette, J.P, Michot, N, Benard, T, Below, P, Vade, I, Chatreaux, F, Lebourg, G, Pilorge, F, Angouillant-Boniface, O, Louboutin, A, Lengauer, C, Schio, L. | | Deposit date: | 2012-02-20 | | Release date: | 2012-05-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Optimization of New Benzimidazole- and Benzoxazole-Pyrimidone Selective Pi3Kbeta Inhibitors for the Treatment of Phosphatase and Tensin Homologue (Pten)-Deficient Cancers.
J.Med.Chem., 55, 2012
|
|
4BFR
 
 | | Discovery and Optimization of Pyrimidone Indoline Amide PI3Kbeta Inhibitors for the Treatment of Phosphatase and TENsin homologue (PTEN)-Deficient Cancers | | Descriptor: | 2-[2-(2-METHYL-2,3-DIHYDRO-INDOL-1-YL)-2-OXO-ETHYL]-6-MORPHOLIN-4-YL-3H-PYRIMIDIN-4-ONE, PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE 3-KINASE CATALYTIC S SUBUNIT BETA ISOFORM | | Authors: | Certal, V, Carry, J.C, Halley, F, Virone-Oddos, A, Thompson, F, Filoche-Romme, B, El-Ahmad, Y, Karlsson, A, Charrier, V, Delorme, C, Rak, A, Abecassis, P.Y, Amara, C, Vincent, L, Bonnevaux, H, Nicolas, J.P, Mathieu, M, Bertrand, T, Marquette, J.P, Michot, N, Benard, T, Perrin, M.A, Perron, S, Monget, S, Gruss-Leleu, F, Doerflinger, G, Guizani, H, Brollo, M, Delbarre, L, Bertin, L, Richepin, P, Loyau, V, Garcia-Echeverria, C, Lengauer, C, Schio, L. | | Deposit date: | 2013-03-22 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Optimization of Pyrimidone Indoline Amide Pi3Kbeta Inhibitors for the Treatment of Phosphatase and Tensin Homologue (Pten)-Deficient Cancers.
J.Med.Chem., 57, 2014
|
|
1AB3
 
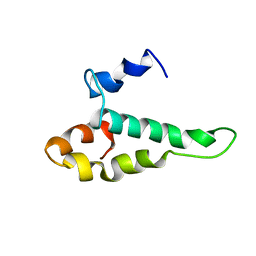 | | RIBOSOMAL PROTEIN S15 FROM THERMUS THERMOPHILUS, NMR, 26 STRUCTURES | | Descriptor: | RIBOSOMAL RNA BINDING PROTEIN S15 | | Authors: | Berglund, H, Rak, A, Serganov, A, Garber, M, Hard, T. | | Deposit date: | 1997-02-03 | | Release date: | 1997-04-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ribosomal RNA binding protein S15 from Thermus thermophilus.
Nat.Struct.Biol., 4, 1997
|
|
3ZLW
 
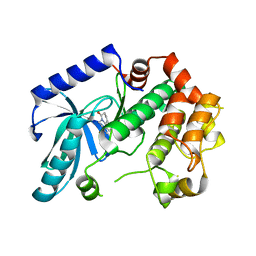 | | Crystal structure of MEK1 in complex with fragment 3 | | Descriptor: | (1R)-1-hydroxy-1-methyl-2,3,6,7-tetrahydro-1H,5H-pyrido[3,2,1-ij]quinolin-5-one, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1 MAPK/ERK KINASE 1, MEK 1, ... | | Authors: | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | Deposit date: | 2013-02-04 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3ZLX
 
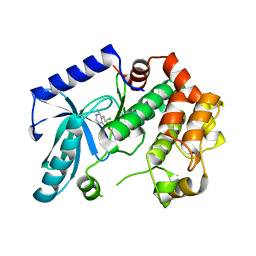 | | Crystal structure of MEK1 in complex with fragment 18 | | Descriptor: | 7-choro-6-[(3R)-pyrrolidin-3-ylmethoxy]isoquinolin-1(2H)-one, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1 | | Authors: | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | Deposit date: | 2013-02-04 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4ASZ
 
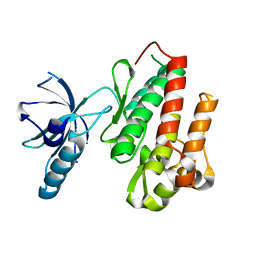 | | Crystal structure of apo TrkB kinase domain | | Descriptor: | BDNF/NT-3 GROWTH FACTORS RECEPTOR | | Authors: | Bertrand, T, Kothe, M, Liu, J, Dupuy, A, Rak, A, Berne, P.F, Davis, S, Gladysheva, T, Valtre, C, Crenne, J.Y, Mathieu, M. | | Deposit date: | 2012-05-03 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structures of Trka and Trkb Suggest Key Regions for Achieving Selective Inhibition.
J.Mol.Biol., 423, 2012
|
|
