7DKT
 
 | |
7DKX
 
 | |
7DKU
 
 | |
7DKW
 
 | |
7DKV
 
 | |
7DKY
 
 | |
7DKS
 
 | |
6AEJ
 
 | | Crystal structure of human FTO in complex with small-molecule inhibitors | | Descriptor: | (E)-3-[3-nitro-4,5-bis(oxidanyl)phenyl]-2-(1,3-oxazinan-3-ylcarbonyl)prop-2-enenitrile, Alpha-ketoglutarate-dependent dioxygenase FTO,Alpha-ketoglutarate-dependent dioxygenase FTO, ZINC ION | | Authors: | Wang, Y, Cao, R, Peng, S, Zhang, W, Huang, N. | | Deposit date: | 2018-08-05 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of entacapone as a chemical inhibitor of FTO mediating metabolic regulation through FOXO1.
Sci Transl Med, 11, 2019
|
|
6AK4
 
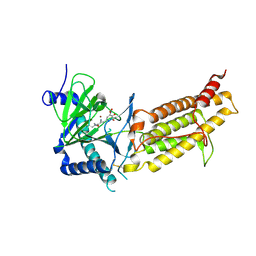 | | Crystal structure of human FTO in complex with small-molecule inhibitors | | Descriptor: | (~{E})-2-cyano-~{N},~{N}-diethyl-3-[3-nitro-4,5-bis(oxidanyl)phenyl]prop-2-enamide, Alpha-ketoglutarate-dependent dioxygenase FTO,Alpha-ketoglutarate-dependent dioxygenase FTO, ZINC ION | | Authors: | Wang, Y, Cao, R, Peng, S, Zhang, W, Huang, N. | | Deposit date: | 2018-08-30 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of entacapone as a chemical inhibitor of FTO mediating metabolic regulation through FOXO1.
Sci Transl Med, 11, 2019
|
|
5UCJ
 
 | | Hsp90b N-terminal domain with inhibitors | | Descriptor: | (5-fluoroisoindolin-2-yl)(4-hydroxy-5-isopropylbenzo[d]isoxazol-7-yl)methanone, DIMETHYL SULFOXIDE, Heat shock protein HSP 90-beta | | Authors: | Peng, S, Balch, M, Matts, R, Deng, J. | | Deposit date: | 2016-12-22 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.693 Å) | | Cite: | Structure-guided design of an Hsp90 beta N-terminal isoform-selective inhibitor.
Nat Commun, 9, 2018
|
|
5UCI
 
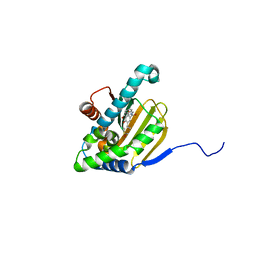 | | Hsp90b N-terminal domain with inhibitors | | Descriptor: | (2,4-Dihydroxy-3-(hydroxymethyl)-5-isopropylphenyl)(isoindolin-2-yl)methanone, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Peng, S, Balch, M, Matts, R, Deng, J. | | Deposit date: | 2016-12-22 | | Release date: | 2018-01-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-guided design of an Hsp90 beta N-terminal isoform-selective inhibitor.
Nat Commun, 9, 2018
|
|
5UCH
 
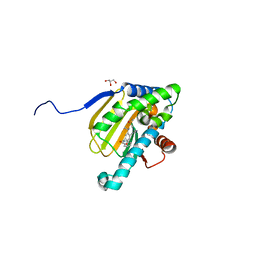 | | Hsp90b N-terminal domain with inhibitors | | Descriptor: | 2-(5-Hydroxy-4-(isoindoline-2-carbonyl)-2-isopropylphenyl)acetonitrile, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Peng, S, Balch, M, Matts, R, Deng, J. | | Deposit date: | 2016-12-22 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.654 Å) | | Cite: | Structure-guided design of an Hsp90 beta N-terminal isoform-selective inhibitor.
Nat Commun, 9, 2018
|
|
7W3Z
 
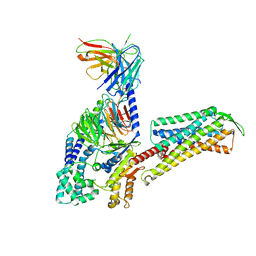 | | Cryo-EM Structure of Human Gastrin Releasing Peptide Receptor in complex with the agonist Gastrin Releasing Peptide and Gq heterotrimers | | Descriptor: | Gastrin Releasing Peptide PRGNHWAVGHLM(NH2), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhan, Y, Peng, S, Zhang, H. | | Deposit date: | 2021-11-26 | | Release date: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of human gastrin-releasing peptide receptors bound to antagonist and agonist for cancer and itch therapy.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7W40
 
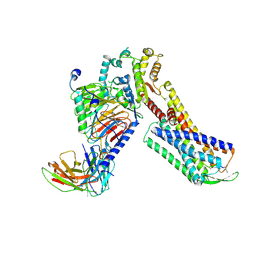 | | Cryo-EM Structure of Human Gastrin Releasing Peptide Receptor in complex with the agonist Bombesin (6-14) [D-Phe6, beta-Ala11, Phe13, Nle14] and Gq heterotrimers | | Descriptor: | Bombesin, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhan, Y, Peng, S, Zhang, H. | | Deposit date: | 2021-11-26 | | Release date: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of human gastrin-releasing peptide receptors bound to antagonist and agonist for cancer and itch therapy.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JBO
 
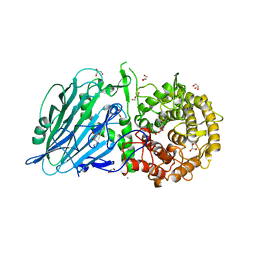 | | Crystal structure of TxGH116 from Thermoanaerobacterium xylanolyticum with isofagomine | | Descriptor: | 1,2-ETHANEDIOL, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, CALCIUM ION, ... | | Authors: | Pengthaisong, S, Ketudat Cairns, J.R. | | Deposit date: | 2023-05-09 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for inhibition of a GH116 beta-glucosidase and its missense mutants by GBA2 inhibitors: Crystallographic and quantum chemical study.
Chem.Biol.Interact., 384, 2023
|
|
7BZM
 
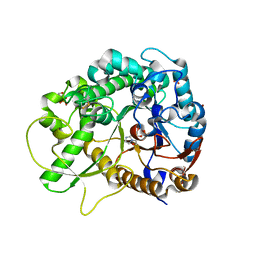 | | Crystal structure of rice Os3BGlu7 with glucoimidazole | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, GLUCOIMIDAZOLE, ... | | Authors: | Pengthaisong, S, Ketudat Cairns, J.R, Tankrathok, A. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Specific Glucoimidazole and Mannoimidazole Binding by Os3BGlu7.
Biomolecules, 10, 2020
|
|
6WIG
 
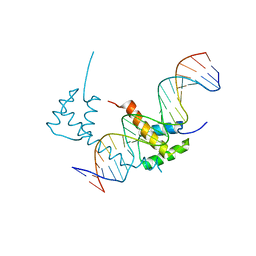 | | Structure of STENOFOLIA Protein HD domain bound with DNA | | Descriptor: | DNA (5'-D(P*CP*TP*TP*GP*AP*AP*TP*AP*AP*AP*TP*CP*AP*TP*TP*AP*AP*TP*TP*TP*GP*C)-3'), DNA (5'-D(P*GP*CP*AP*AP*AP*TP*TP*AP*AP*TP*GP*AP*TP*TP*TP*AP*TP*TP*CP*AP*AP*G)-3'), STENOFOLIA | | Authors: | Deng, J, Peng, S, Pathak, P. | | Deposit date: | 2020-04-09 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the unique tetrameric STENOFOLIA homeodomain bound with target promoter DNA.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7LSZ
 
 | | Hsp90a N-terminal inhibitor | | Descriptor: | Heat shock protein HSP 90-alpha, {3-[(2,3-dihydro-1,4-benzodioxin-6-yl)sulfanyl]-4-hydroxyphenyl}(1,3-dihydro-2H-isoindol-2-yl)methanone | | Authors: | Balch, M, Peng, S, Deng, J, Matts, R. | | Deposit date: | 2021-02-18 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Selective Inhibition of the Hsp90 alpha Isoform.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7LT0
 
 | | Hsp90a N-terminal inhibitor | | Descriptor: | (1,3-dihydro-2H-isoindol-2-yl){3-[(3,4-dimethylphenyl)sulfanyl]-4-hydroxyphenyl}methanone, Heat shock protein HSP 90-alpha | | Authors: | Balch, M, Peng, S, Deng, J, Matts, R. | | Deposit date: | 2021-02-18 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Selective Inhibition of the Hsp90 alpha Isoform.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6BR9
 
 | | Structure of A6 reveals a novel lipid transporter | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, Protein A6 homolog, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Deng, J, Peng, S, Pathak, P. | | Deposit date: | 2017-11-30 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a lipid-bound viral membrane assembly protein reveals a modality for enclosing the lipid bilayer.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BR8
 
 | | Structure of A6 reveals a novel lipid transporter | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, Protein A6 homolog, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Deng, J, Peng, S, Pathak, P. | | Deposit date: | 2017-11-30 | | Release date: | 2018-06-20 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a lipid-bound viral membrane assembly protein reveals a modality for enclosing the lipid bilayer.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5UC4
 
 | | Hsp90b N-terminal domain with inhibitors | | Descriptor: | 5-Hydroxy-4-(isoindoline-2-carbonyl)-2-isopropylbenzaldehyde, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Deng, J, Peng, S, Balch, M, Matts, R. | | Deposit date: | 2016-12-21 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-guided design of an Hsp90 beta N-terminal isoform-selective inhibitor.
Nat Commun, 9, 2018
|
|
7U8W
 
 | | hTRAP1 with inhibitors | | Descriptor: | Heat shock protein 75 kDa, mitochondrial, [5-(4-fluoro-2H-isoindole-2-carbonyl)-2-hydroxyphenyl](5-fluoro-2H-isoindol-2-yl)methanone | | Authors: | Deng, J, Matts, R, Peng, S. | | Deposit date: | 2022-03-09 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.706 Å) | | Cite: | Elucidation of novel TRAP1-Selective inhibitors that regulate mitochondrial processes.
Eur.J.Med.Chem., 258, 2023
|
|
7U8X
 
 | | hTRAP1 with inhibitors | | Descriptor: | Heat shock protein 75 kDa, mitochondrial, [5-(4-fluoro-2H-isoindole-2-carbonyl)-2-hydroxyphenyl](5-methoxy-2H-isoindol-2-yl)methanone | | Authors: | Deng, J, Matts, R, Peng, S. | | Deposit date: | 2022-03-09 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Elucidation of novel TRAP1-Selective inhibitors that regulate mitochondrial processes.
Eur.J.Med.Chem., 258, 2023
|
|
7U8V
 
 | | hTRAP1 with inhibitors | | Descriptor: | (4-hydroxy-1,3-phenylene)bis[(2H-isoindol-2-yl)methanone], Heat shock protein 75 kDa, mitochondrial | | Authors: | Deng, J, Matts, R, Peng, S. | | Deposit date: | 2022-03-09 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.448 Å) | | Cite: | Elucidation of novel TRAP1-Selective inhibitors that regulate mitochondrial processes.
Eur.J.Med.Chem., 258, 2023
|
|
