3M45
 
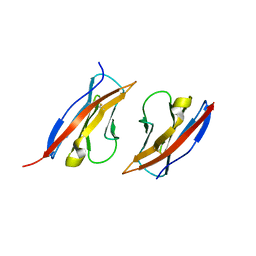 | | Crystal structure of Ig1 domain of mouse SynCAM 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cell adhesion molecule 2 | | Authors: | Yue, L, Modis, Y. | | Deposit date: | 2010-03-10 | | Release date: | 2010-09-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | N-glycosylation at the SynCAM (synaptic cell adhesion molecule) immunoglobulin interface modulates synaptic adhesion.
J.Biol.Chem., 285, 2010
|
|
5VSL
 
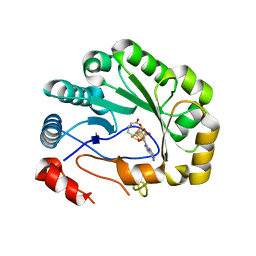 | | Crystal structure of viperin with bound [4Fe-4S] cluster and S-adenosylhomocysteine (SAH) | | Descriptor: | IRON/SULFUR CLUSTER, Radical S-adenosyl methionine domain-containing protein 2, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Fenwick, M.K, Li, Y, Cresswell, P, Modis, Y, Ealick, S.E. | | Deposit date: | 2017-05-11 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.972 Å) | | Cite: | Structural studies of viperin, an antiviral radical SAM enzyme.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VSM
 
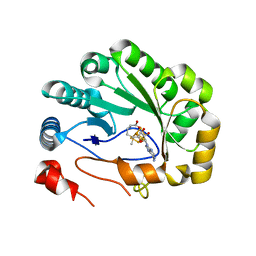 | | Crystal structure of viperin with bound [4Fe-4S] cluster, 5'-deoxyadenosine, and L-methionine | | Descriptor: | 5'-DEOXYADENOSINE, IRON/SULFUR CLUSTER, METHIONINE, ... | | Authors: | Fenwick, M.K, Li, Y, Cresswell, P, Modis, Y, Ealick, S.E. | | Deposit date: | 2017-05-11 | | Release date: | 2017-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural studies of viperin, an antiviral radical SAM enzyme.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
9F3P
 
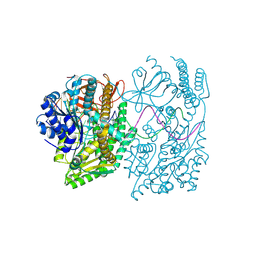 | | Cryo-EM structure of the A946T MDA5-dsRNA filament | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, RNA (5'-R(P*GP*UP*CP*AP*AP*GP*CP*CP*GP*AP*GP*GP*AP*GP*A)-3'), RNA (5'-R(P*UP*CP*UP*CP*CP*UP*CP*GP*GP*CP*UP*UP*GP*AP*C)-3'), ... | | Authors: | Singh, R, Herrero del Valle, A, Modis, Y. | | Deposit date: | 2024-04-25 | | Release date: | 2025-02-26 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Molecular basis of autoimmune disease protection by MDA5 variants.
Cell Rep, 44, 2025
|
|
9F2W
 
 | | Cryo-EM structure of the I923V MDA5-dsRNA filament in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Interferon-induced helicase C domain-containing protein 1, RNA (5'-R(P*AP*UP*CP*UP*CP*CP*UP*CP*GP*GP*CP*UP*UP*G)-3'), ... | | Authors: | Singh, R, Herrero del Valle, A, Modis, Y. | | Deposit date: | 2024-04-24 | | Release date: | 2025-02-26 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (4.21 Å) | | Cite: | Molecular basis of autoimmune disease protection by MDA5 variants.
Cell Rep, 44, 2025
|
|
9F2L
 
 | | Cryo-EM structure of the I923V MDA5-dsRNA filament with ADP-AlF4 bound and 73-degree helical twist | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Interferon-induced helicase C domain-containing protein 1, RNA (5'-R(P*CP*AP*AP*GP*CP*CP*GP*AP*GP*GP*AP*GP*AP*G)-3'), ... | | Authors: | Singh, R, Herrero del Valle, A, Modis, Y. | | Deposit date: | 2024-04-23 | | Release date: | 2025-02-26 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Molecular basis of autoimmune disease protection by MDA5 variants.
Cell Rep, 44, 2025
|
|
9F20
 
 | | Cryo-EM structure of the I923V MDA5-dsRNA filament with ADP-AlF4 bound and 88-degree helical twist | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Interferon-induced helicase C domain-containing protein 1, RNA (5'-R(P*GP*UP*CP*AP*AP*GP*CP*CP*GP*AP*GP*GP*AP*GP*A)-3'), ... | | Authors: | Singh, R, Herrero del Valle, A, Modis, Y. | | Deposit date: | 2024-04-20 | | Release date: | 2025-02-26 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (4.13 Å) | | Cite: | Molecular basis of autoimmune disease protection by MDA5 variants.
Cell Rep, 44, 2025
|
|
9F1U
 
 | | Cryo-EM structure of the I923V MDA5-dsRNA filament with ADP-AlF4 bound and 81-degree helical twist | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Interferon-induced helicase C domain-containing protein 1, RNA (5'-R(P*AP*UP*CP*UP*CP*CP*UP*CP*GP*GP*CP*UP*UP*G)-3'), ... | | Authors: | Singh, R, Herrero del Valle, A, Modis, Y. | | Deposit date: | 2024-04-20 | | Release date: | 2025-02-26 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Molecular basis of autoimmune disease protection by MDA5 variants.
Cell Rep, 44, 2025
|
|
9F0J
 
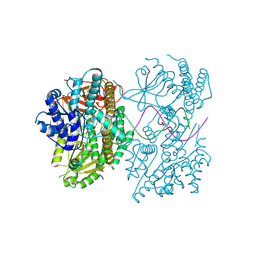 | | Cryo-EM structure of the I923V MDA5-dsRNA filament without nucleotide | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, RNA (5'-CGUCAUGCGCAUGGA-3'), RNA (5'-UCCAUGCGCAUGACG-3'), ... | | Authors: | Singh, R, Herrero del Valle, A, Modis, Y. | | Deposit date: | 2024-04-16 | | Release date: | 2025-02-26 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Molecular basis of autoimmune disease protection by MDA5 variants.
Cell Rep, 44, 2025
|
|
3NEC
 
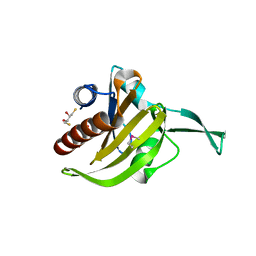 | | Crystal Structure of Toxoplasma gondii Profilin | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, Inflammatory profilin, SULFATE ION | | Authors: | Kucera, K, Modis, Y. | | Deposit date: | 2010-06-08 | | Release date: | 2010-09-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based analysis of Toxoplasma gondii profilin: a parasite-specific motif is required for recognition by Toll-like receptor 11.
J.Mol.Biol., 403, 2010
|
|
8P0E
 
 | |
8P0C
 
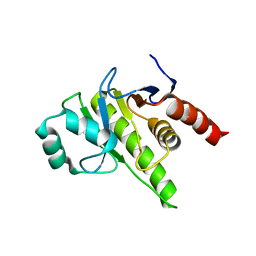 | | Rubella virus p150 macro domain (apo) | | Descriptor: | Non-structural polyprotein p200 | | Authors: | Stoll, G.A, Modis, Y. | | Deposit date: | 2023-05-10 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure and biochemical activity of the macrodomain from rubella virus p150.
J.Virol., 98, 2024
|
|
8QFB
 
 | |
6T1B
 
 | |
6T0Y
 
 | |
7NIQ
 
 | |
7NIC
 
 | |
7NGA
 
 | |
1RH5
 
 | | The structure of a protein conducting channel | | Descriptor: | Preprotein translocase secE subunit, Preprotein translocase secY subunit, SecBeta | | Authors: | van den Berg, B, Clemons Jr, W.M, Collinson, I, Modis, Y, Hartmann, E, Harrison, S.C, Rapoport, T.A. | | Deposit date: | 2003-11-13 | | Release date: | 2004-01-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray structure of a protein-conducting channel
Nature, 427, 2004
|
|
7BKQ
 
 | | CryoEM structure of MDA5-dsRNA filament in complex with ADP with 92-degree helical twist | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Isoform 2 of Interferon-induced helicase C domain-containing protein 1, RNA (5'-R(P*CP*GP*UP*CP*AP*UP*GP*CP*GP*CP*AP*UP*GP*GP*A)-3'), ... | | Authors: | Yu, Q, Modis, Y. | | Deposit date: | 2021-01-17 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | MDA5 disease variant M854K prevents ATP-dependent structural discrimination of viral and cellular RNA.
Nat Commun, 12, 2021
|
|
7BKP
 
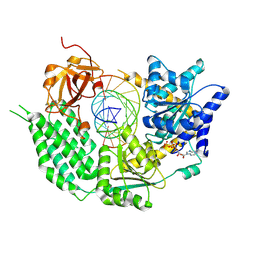 | | CryoEM structure of disease related M854K MDA5-dsRNA filament in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Interferon-induced helicase C domain-containing protein 1, MAGNESIUM ION, ... | | Authors: | Singh, R, Herrero del Valle, A, Yu, Q, Modis, Y. | | Deposit date: | 2021-01-16 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | MDA5 disease variant M854K prevents ATP-dependent structural discrimination of viral and cellular RNA.
Nat Commun, 12, 2021
|
|
1RHZ
 
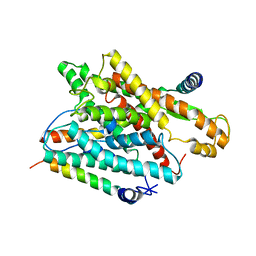 | | The structure of a protein conducting channel | | Descriptor: | Preprotein translocase secE subunit, Preprotein translocase secY subunit, SecBeta | | Authors: | van den Berg, B, Clemons Jr, W.M, Collinson, I, Modis, Y, Hartmann, E, Harrison, S.C, Rapoport, T.A. | | Deposit date: | 2003-11-15 | | Release date: | 2004-01-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | X-ray structure of a protein-conducting channel.
Nature, 427, 2004
|
|
4JNT
 
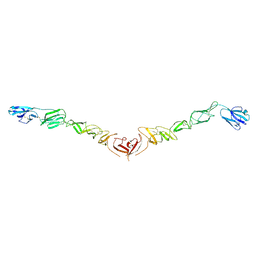 | |
4HJ1
 
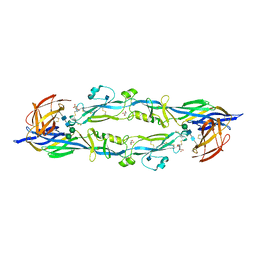 | |
4HJC
 
 | |
