9B98
 
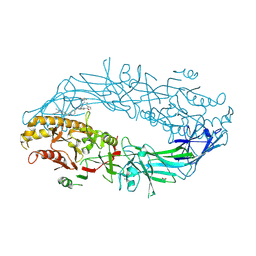 | | Crystal structure of the human PAD2 protein bound to small molecule | | Descriptor: | (5P)-N,N-diethyl-2-fluoro-5-(2-[({1-[2-(methylamino)-2-oxoethyl]cyclohexyl}methyl)amino]-6-{methyl[1-(2-methyl-1-phenyl-1H-1,3-benzimidazole-5-carbonyl)piperidin-4-yl]amino}pyrimidin-4-yl)benzamide, ACETATE ION, CALCIUM ION, ... | | Authors: | Byrnes, L.J, Vajdos, F. | | Deposit date: | 2024-04-01 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Discovery, Characterization, and Structure of a Cell Active PAD2 Inhibitor Acting through a Novel Allosteric Mechanism.
Acs Chem.Biol., 19, 2024
|
|
4ZO5
 
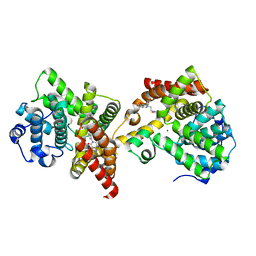 | | PDE10 complexed with 4-isopropoxy-2-(2-(3-(4-methoxyphenyl)-4-oxo-3,4-dihydroquinazolin-2-yl)ethyl)isoindoline-1,3-dione | | Descriptor: | 2-{2-[3-(4-methoxyphenyl)-4-oxo-3,4-dihydroquinazolin-2-yl]ethyl}-4-(propan-2-yloxy)-1H-isoindole-1,3(2H)-dione, 3-(1-hydroxy-2-methylpropan-2-yl)-5-phenyl-3,5-dihydro-1H-imidazo[4,5-c][1,8]naphthyridine-2,4-dione, MAGNESIUM ION, ... | | Authors: | Yan, Y. | | Deposit date: | 2015-05-06 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of [(11)C]MK-8193 as a PET tracer to measure target engagement of phosphodiesterase 10A (PDE10A) inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
9M3N
 
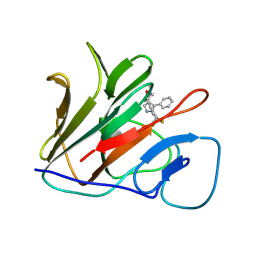 | | Crystal structure of human TRIM21 PRYSPRY in complex with T-02 | | Descriptor: | 1-ethyl-~{N}-methyl-5-phenyl-~{N}-[3-[3-(trifluoromethyl)phenyl]cyclobutyl]pyrazole-4-carboxamide, E3 ubiquitin-protein ligase TRIM21 | | Authors: | Zhang, L.J, Zhang, L.Y. | | Deposit date: | 2025-03-03 | | Release date: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | TRIM21-Driven Degradation of BRD4: Development of Heterobifunctional Degraders and Investigation of Recruitment and Selectivity Mechanisms.
J.Chem.Inf.Model., 2025
|
|
7BAJ
 
 | |
7BAL
 
 | |
7BAK
 
 | |
5IMX
 
 | |
5JLJ
 
 | | Crystal Structure of KPT8602 in complex with CRM1-Ran-RanBP1 | | Descriptor: | (2R)-3-{3-[3,5-bis(trifluoromethyl)phenyl]-1H-1,2,4-triazol-1-yl}-2-(pyrimidin-5-yl)propanamide, CHLORIDE ION, Exportin-1, ... | | Authors: | Fung, H.Y, Chook, Y.M. | | Deposit date: | 2016-04-27 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Next-generation XPO1 inhibitor shows improved efficacy and in vivo tolerability in hematological malignancies.
Leukemia, 30, 2016
|
|
6XFN
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with UAW243 | | Descriptor: | 3C-like proteinase, GLYCEROL, UAW243 | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-15 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
6XBI
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAW248 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-06 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
6XBH
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAW247 | | Descriptor: | 3C-like proteinase, GLYCEROL, SODIUM ION, ... | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-06 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
6XA4
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with UAW241 | | Descriptor: | 3C-like proteinase, GLYCEROL, inhibitor UAW241 | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
7DTT
 
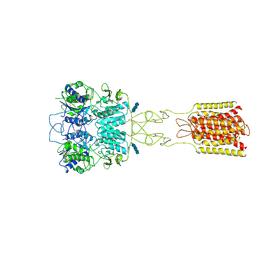 | | Human Calcium-Sensing Receptor bound with calcium ions | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ling, S.L, Tian, C.L, Shi, P, Liu, S.L, Meng, X.Y, Sun, D.M, Liu, L, Shi, C.W. | | Deposit date: | 2021-01-06 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural mechanism of cooperative activation of the human calcium-sensing receptor by Ca 2+ ions and L-tryptophan.
Cell Res., 31, 2021
|
|
7DTW
 
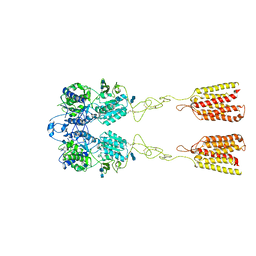 | | Human Calcium-Sensing Receptor in the inactive close-close conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Extracellular calcium-sensing receptor | | Authors: | Ling, S.L, Tian, C.L, Shi, P, Liu, S.L, Meng, X.Y, Liu, L, Sun, D.M, Shi, C.W. | | Deposit date: | 2021-01-06 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural mechanism of cooperative activation of the human calcium-sensing receptor by Ca 2+ ions and L-tryptophan.
Cell Res., 31, 2021
|
|
7DTV
 
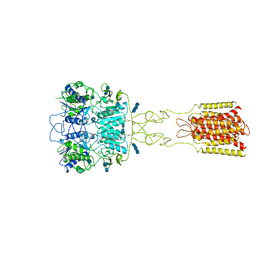 | | Human Calcium-Sensing Receptor bound with L-Trp and calcium ions | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ling, S.L, Tian, C.L, Shi, P, Liu, S.L, Meng, X.Y, Liu, L, Sun, D.M, Shi, C.W. | | Deposit date: | 2021-01-06 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural mechanism of cooperative activation of the human calcium-sensing receptor by Ca 2+ ions and L-tryptophan.
Cell Res., 31, 2021
|
|
7DTU
 
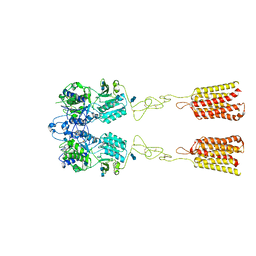 | | Human Calcium-Sensing Receptor bound with L-Trp | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Extracellular calcium-sensing receptor, ... | | Authors: | Ling, S.L, Tian, C.L, Shi, P, Liu, S.L, Meng, X.Y, Liu, L, Sun, D.M, Shi, C.W. | | Deposit date: | 2021-01-06 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural mechanism of cooperative activation of the human calcium-sensing receptor by Ca 2+ ions and L-tryptophan.
Cell Res., 31, 2021
|
|
6XBG
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAW246 | | Descriptor: | 3C-like proteinase, GLYCEROL, SODIUM ION, ... | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-05 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
7RN0
 
 | |
7RN1
 
 | |
5USL
 
 | | Structure of vaccinia virus D8 protein bound to human Fab vv304 | | Descriptor: | Fab vv304 Heavy chain, Fab vv304 Light chain, IMV membrane protein | | Authors: | Zajonc, D.M. | | Deposit date: | 2017-02-13 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-function characterization of three human antibodies targeting the vaccinia virus adhesion molecule D8.
J. Biol. Chem., 293, 2018
|
|
5USH
 
 | | Structure of vaccinia virus D8 protein bound to human Fab vv66 | | Descriptor: | Fab vv66 heavy chain, Fab vv66 light chain, IMV membrane protein | | Authors: | Zajonc, D.M. | | Deposit date: | 2017-02-13 | | Release date: | 2017-09-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-function characterization of three human antibodies targeting the vaccinia virus adhesion molecule D8.
J. Biol. Chem., 293, 2018
|
|
6NCU
 
 | | Interleukin-37 residues 53-206- dimer | | Descriptor: | Interleukin-37 | | Authors: | Eisenmesser, E.Z. | | Deposit date: | 2018-12-12 | | Release date: | 2019-03-13 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Interleukin-37 monomer is the active form for reducing innate immunity.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
8CZ9
 
 | |
9II5
 
 | | Crystal structure of human TRIM21 PRYSPRY in complex with compound 1 | | Descriptor: | E3 ubiquitin-protein ligase TRIM21, ~{N}-[(1-fluoranylcyclohexyl)methyl]-~{N}-methyl-4-(2-methylsulfanylphenyl)-2-methylsulfonyl-benzamide | | Authors: | Zhang, L.Y. | | Deposit date: | 2024-06-19 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Chemically Induced Nuclear Pore Complex Protein Degradation via TRIM21.
Acs Chem.Biol., 20, 2025
|
|
7JRN
 
 | | Crystal structure of the wild type SARS-CoV-2 papain-like protease (PLPro) with inhibitor GRL0617 | | Descriptor: | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, Non-structural protein 3, SULFATE ION, ... | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-08-12 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of SARS-CoV-2 Papain-like Protease Inhibitors through a Combination of High-Throughput Screening and a FlipGFP-Based Reporter Assay.
Acs Cent.Sci., 7, 2021
|
|
