6Z98
 
 | | NMR solution structure of the peach allergen Pru p 1.0101 | | Descriptor: | Major allergen Pru p 1 | | Authors: | Eidelpes, R, Fuehrer, S, Hofer, F, Kamenik, A.S, Liedl, K.R, Tollinger, M. | | Deposit date: | 2020-06-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Zeatin Binding of the Peach Allergen Pru p 1 .
J.Agric.Food Chem., 69, 2021
|
|
2JNR
 
 | | Discovery and optimization of a natural HIV-1 entry inhibitor targeting the gp41 fusion peptide | | Descriptor: | ENV polyprotein, VIR165 | | Authors: | Munch, J, Standker, L, Adermann, K, Schulz, A, Pohlmann, S, Chaipan, C, Biet, T, Peters, T, Meyer, B, Wilhelm, D, Lu, H, Jing, W, Jiang, S, Forssmann, W, Kirchhoff, F. | | Deposit date: | 2007-02-01 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Discovery and Optimization of a Natural HIV-1 Entry Inhibitor Targeting the gp41 Fusion Peptide.
Cell(Cambridge,Mass.), 129, 2007
|
|
4PRU
 
 | | Crystal structure of dimethyllysine hen egg-white lysozyme in complex with sclx4 at 2.2 A resolution | | Descriptor: | 25,26,27,28-tetrahydroxypentacyclo[19.3.1.1~3,7~.1~9,13~.1~15,19~]octacosa-1(25),3(28),4,6,9(27),10,12,15(26),16,18,21,23-dodecaene-5,11,17,23-tetrasulfonic acid, GLYCEROL, Lysozyme C | | Authors: | McGovern, R.E, Lyons, J.A, Crowley, P.B. | | Deposit date: | 2014-03-06 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural study of a small molecule receptor bound to dimethyllysine in lysozyme.
Chem Sci, 6, 2015
|
|
4N0J
 
 | | Crystal structure of dimethyllysine hen egg-white lysozyme in complex with sclx4 at 1.9 A resolution | | Descriptor: | 25,26,27,28-tetrahydroxypentacyclo[19.3.1.1~3,7~.1~9,13~.1~15,19~]octacosa-1(25),3(28),4,6,9(27),10,12,15(26),16,18,21,23-dodecaene-5,11,17,23-tetrasulfonic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | McGovern, R.E, Crowley, P.B. | | Deposit date: | 2013-10-02 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural study of a small molecule receptor bound to dimethyllysine in lysozyme.
Chem Sci, 6, 2015
|
|
2WND
 
 | | Structure of an S100A7 triple mutant | | Descriptor: | CALCIUM ION, PROTEIN S100-A7, ZINC ION | | Authors: | West, N.R, Farnell, B, Watson, P.H, Boulanger, M.J. | | Deposit date: | 2009-07-08 | | Release date: | 2009-11-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Characterization of a Triple Mutant Form of S100A7 Defective for Jab1 Binding.
Protein Sci., 18, 2009
|
|
8OK1
 
 | |
8OKF
 
 | | WD repeat containing protein 5 (WDR5)- PER2 peptide | | Descriptor: | CHLORIDE ION, Glutathione S-transferase class-mu 26 kDa isozyme,WD repeat domain 5, PENTAETHYLENE GLYCOL, ... | | Authors: | Wolf, E, Boergel, A. | | Deposit date: | 2023-03-28 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A structural competition involving WDR5 times circadian oscillations
To Be Published
|
|
5FA5
 
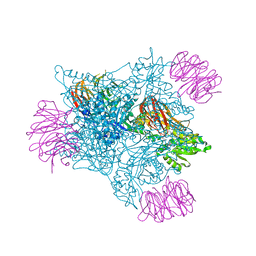 | |
6TRK
 
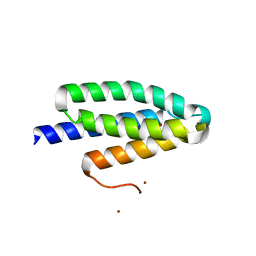 | | Phl p 6 fold stabilized mutant - S46Y | | Descriptor: | Pollen allergen Phl p 6, ZINC ION | | Authors: | Soh, W.T, Brandstetter, H. | | Deposit date: | 2019-12-19 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | In silico Design of Phl p 6 Variants With Altered Fold-Stability Significantly Impacts Antigen Processing, Immunogenicity and Immune Polarization.
Front Immunol, 11, 2020
|
|
3KRR
 
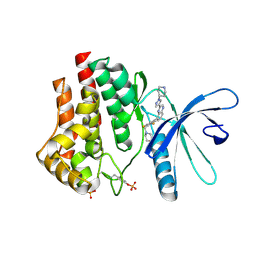 | | Crystal Structure of JAK2 complexed with a potent quinoxaline ATP site inhibitor | | Descriptor: | 8-[3,5-difluoro-4-(morpholin-4-ylmethyl)phenyl]-2-(1-piperidin-4-yl-1H-pyrazol-4-yl)quinoxaline, Tyrosine-protein kinase JAK2 | | Authors: | Tavares, G.A, Gerspacher, M, Kroemer, M, Scheufler, C. | | Deposit date: | 2009-11-19 | | Release date: | 2010-07-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Potent and Selective Inhibition of Polycythemia by the Quinoxaline JAK2 Inhibitor NVP-BSK805
Mol.Cancer Ther., 9, 2010
|
|
1RAX
 
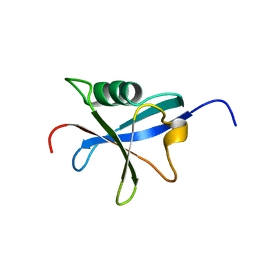 | |
2N0X
 
 | | Three dimensional structure of EPI-X4, a human albumin-derived peptide that regulates innate immunity through the CXCR4/CXCL12 chemotactic axis and antagonizes HIV-1 entry | | Descriptor: | Serum albumin | | Authors: | Perez-Castells, J, Canales, A, Jimenez-Barbero, J, Gimenez-Gallego, G. | | Deposit date: | 2015-03-18 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Discovery and characterization of an endogenous CXCR4 antagonist.
Cell Rep, 11, 2015
|
|
3UGC
 
 | | Structural basis of Jak2 inhibition by the type II inhibtor NVP-BBT594 | | Descriptor: | 5-{[6-(acetylamino)pyrimidin-4-yl]oxy}-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}-2,3-dihydro-1H-indole-1-carboxamide, MALONATE ION, Tyrosine-protein kinase JAK2 | | Authors: | Scheufler, C, Tavares, G.A, Manley, P.W, Pissot-Soldermann, C, Kroemer, M. | | Deposit date: | 2011-11-02 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Modulation of activation-loop phosphorylation by JAK inhibitors is binding mode dependent.
Cancer Discov, 2, 2012
|
|
2RGF
 
 | | RBD OF RAL GUANOSINE-NUCLEOTIDE EXCHANGE FACTOR (PROTEIN), NMR, 10 STRUCTURES | | Descriptor: | RALGEF-RBD | | Authors: | Geyer, M, Herrmann, C, Wittinghofer, A, Kalbitzer, H.R. | | Deposit date: | 1997-02-13 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Ras-binding domain of RalGEF and implications for Ras binding and signalling.
Nat.Struct.Biol., 4, 1997
|
|
7C53
 
 | |
4OKF
 
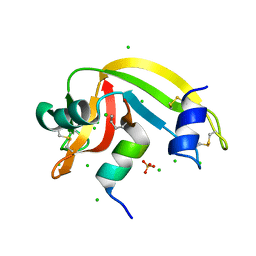 | | RNase S in complex with an artificial peptide | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, SULFATE ION | | Authors: | Genz, M, Strater, N. | | Deposit date: | 2014-01-22 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | X-ray structure of a RNase S variant in complex with an artificial peptide
To be Published
|
|
6UTB
 
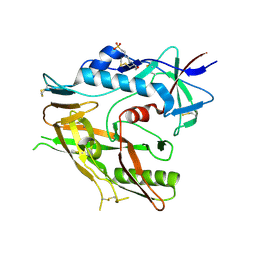 | |
6USW
 
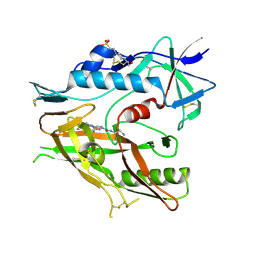 | | CRYSTAL STRUCTURE OF HIV-1 LM/HS CLADE A/E CRF01 GP120 CORE IN COMPLEX WITH (S)-MCG-IV-210 | | Descriptor: | (3S)-N~1~-(2-aminoethyl)-N~3~-(4-chloro-3-fluorophenyl)piperidine-1,3-dicarboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tolbert, W.D, Sherburn, R, Pazgier, M. | | Deposit date: | 2019-10-28 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The HIV-1 Env gp120 Inner Domain Shapes the Phe43 Cavity and the CD4 Binding Site.
Mbio, 11, 2020
|
|
6UT1
 
 | | CRYSTAL STRUCTURE OF HIV-1 LM/HS CLADE A/E CRF01 GP120 CORE IN COMPLEX WITH BNM-III-170 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIV-1 LM/HS clade A/E CRF01 gp120 core, ... | | Authors: | Tolbert, W.D, Sherburn, R, Pazgier, M. | | Deposit date: | 2019-10-29 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The HIV-1 Env gp120 Inner Domain Shapes the Phe43 Cavity and the CD4 Binding Site.
Mbio, 11, 2020
|
|
6UTD
 
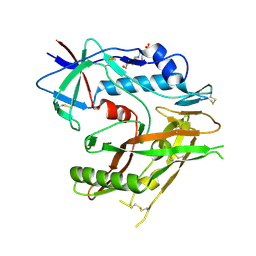 | |
5MO4
 
 | |
121P
 
 | | STRUKTUR UND GUANOSINTRIPHOSPHAT-HYDROLYSEMECHANISMUS DES C-TERMINAL VERKUERZTEN MENSCHLICHEN KREBSPROTEINS P21-H-RAS | | Descriptor: | H-RAS P21 PROTEIN, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Krengel, U, Scheffzek, K, Scherer, A, Kabsch, W, Wittinghofer, A, Pai, E.F. | | Deposit date: | 1991-06-06 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Struktur Und Guanosintriphosphat-Hydrolysemechanismus Des C-Terminal Verkuerzten Menschlichen Krebsproteins P21-H-Ras
Thesis, 1991
|
|
6CM9
 
 | | Structure of the cargo bound AP-1:Arf1:tetherin-Nef closed trimer monomeric subunit | | Descriptor: | ADP-ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Morris, K.L, Buffalo, C.Z, Ren, X, Hurley, J.H. | | Deposit date: | 2018-03-03 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | HIV-1 Nefs Are Cargo-Sensitive AP-1 Trimerization Switches in Tetherin Downregulation.
Cell, 174, 2018
|
|
6CRI
 
 | | Structure of the cargo bound AP-1:Arf1:tetherin-Nef stable closed trimer | | Descriptor: | ADP-ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Morris, K.L, Buffalo, C.Z, Hurley, J.H. | | Deposit date: | 2018-03-18 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | HIV-1 Nefs Are Cargo-Sensitive AP-1 Trimerization Switches in Tetherin Downregulation.
Cell, 174, 2018
|
|
6DFF
 
 | | Structure of the cargo bound AP-1:Arf1:tetherin-Nef monomer | | Descriptor: | ADP-ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Morris, K.L, Buffalo, C.Z, Ren, X, Hurley, J.H. | | Deposit date: | 2018-05-14 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | HIV-1 Nefs Are Cargo-Sensitive AP-1 Trimerization Switches in Tetherin Downregulation.
Cell, 174, 2018
|
|
