2HD6
 
 | | Crystal structure of the human carbonic anhydrase II in complex with a hypoxia-activatable sulfonamide. | | Descriptor: | CHLORIDE ION, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | De Simone, G, Vitale, R.M, Di Fiore, A, Pedone, C. | | Deposit date: | 2006-06-20 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Carbonic anhydrase inhibitors: Hypoxia-activatable sulfonamides incorporating disulfide bonds that target the tumor-associated isoform IX.
J.Med.Chem., 49, 2006
|
|
2HL4
 
 | | Crystal structure analysis of human carbonic anhydrase II in complex with a benzenesulfonamide derivative | | Descriptor: | CHLORIDE ION, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Di Fiore, A, Supuran, C.T, Winum, J.-Y, Montero, J.-L, Pedone, C, Scozzafava, A, De Simone, G. | | Deposit date: | 2006-07-06 | | Release date: | 2007-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Carbonic anhydrase inhibitors: binding of an antiglaucoma glycosyl-sulfanilamide derivative to human isoform II and its consequences for the drug design of enzyme inhibitors incorporating sugar moieties
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2EWB
 
 | | The crystal structure of Bovine Lens Leucine Aminopeptidase in complex with zofenoprilat | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CARBONATE ION, Cytosol aminopeptidase, ... | | Authors: | Alterio, V, Pedone, C, De Simone, G. | | Deposit date: | 2005-11-02 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Metal Ion Substitution in the Catalytic Site Greatly Affects the Binding of Sulfhydryl-Containing Compounds to Leucyl Aminopeptidase.
Biochemistry, 45, 2006
|
|
2F14
 
 | | Tne Crystal Structure of the Human Carbonic Anhydrase II in Complex with a Fluorescent Inhibitor | | Descriptor: | 4-(HYDROXYMERCURY)BENZOIC ACID, 5-{[({2-[4-(AMINOSULFONYL)PHENYL]ETHYL}AMINO)CARBONOTHIOYL]AMINO}-2-(6-HYDROXY-3-OXO-3H-XANTHEN-9-YL)BENZOIC ACID, Carbonic anhydrase 2, ... | | Authors: | Alterio, V, Pedone, C, De Simone, G. | | Deposit date: | 2005-11-14 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Carbonic anhydrase inhibitors: X-ray and molecular modeling study for the interaction of a fluorescent antitumor sulfonamide with isozyme II and IX.
J.Am.Chem.Soc., 128, 2006
|
|
1NO9
 
 | | Design of weakly basic thrombin inhibitors incorporating novel P1 binding functions: molecular and X-ray crystallographic studies. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha Thrombin, N4-(N,N-DIPHENYLCARBAMOYL)-AMINOGUANIDINE, ... | | Authors: | De Simone, G, Menchise, V, Omaggio, S, Pedone, C, Scozzafava, A, Supuran, C.T. | | Deposit date: | 2003-01-16 | | Release date: | 2003-08-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DESIGN OF WEAKLY BASIC THROMBIN INHIBITORS INCORPORATING NOVEL P1 BINDING FUNCTIONS: MOLECULAR AND X-RAY CRYSTALLOGRAPHIC STUDIES
Biochemistry, 42, 2003
|
|
3CZV
 
 | | Crystal structure of the human carbonic anhydrase XIII in complex with acetazolamide | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase 13, GLYCEROL, ... | | Authors: | Di Fiore, A, De Simone, G. | | Deposit date: | 2008-04-30 | | Release date: | 2008-07-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human carbonic anhydrase XIII and its complex with the inhibitor acetazolamide.
Proteins, 74, 2008
|
|
5I69
 
 | | MBP-MamC magnetite-interaction component mutant-D70A | | Descriptor: | Maltose-binding periplasmic protein,Tightly bound bacterial magnetic particle protein,Maltose-binding periplasmic protein, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Nudelman, H, Tercedor, C.V, Kolusheva, S, Gonzalez, T.P, Widdrat, M, Grimberg, N, Levi, H, Nelkenbaum, O, Davidove, G, Faivre, D, Jimenez-Lopez, C, Zarivach, R. | | Deposit date: | 2016-02-16 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-function studies of the magnetite-biomineralizing magnetosome-associated protein MamC.
J.Struct.Biol., 194, 2016
|
|
6T29
 
 | | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) bound to compound 18 (CS587) | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3~{S})-3-azanylpiperidin-1-yl]-4-[[3,5-bis(2-cyanopropan-2-yl)phenyl]amino]pyrimidine-5-carboxamide, Calcium/calmodulin-dependent protein kinase type 1D, ... | | Authors: | Kraemer, A, Sorrell, F, Butterworth, S, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-08 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.484 Å) | | Cite: | Discovery of Highly Selective Inhibitors of Calmodulin-Dependent Kinases That Restore Insulin Sensitivity in the Diet-Induced Obesityin VivoMouse Model.
J.Med.Chem., 63, 2020
|
|
6T28
 
 | | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) bound to compound 19 (CS640) | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3~{S})-3-azanylpiperidin-1-yl]-4-[[2,6-di(propan-2-yl)pyridin-4-yl]amino]pyrimidine-5-carboxamide, Calcium/calmodulin-dependent protein kinase type 1D, ... | | Authors: | Kraemer, A, Sorrell, F, Butterworth, S, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-08 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Highly Selective Inhibitors of Calmodulin-Dependent Kinases That Restore Insulin Sensitivity in the Diet-Induced Obesityin VivoMouse Model.
J.Med.Chem., 63, 2020
|
|
6S1B
 
 | | Crystal Structure of DYRK1A with small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, SULFATE ION, ... | | Authors: | Sorrell, F.J, Henderson, S.H, Redondo, C, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Elkins, J.M. | | Deposit date: | 2019-06-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Kinase Scaffold Repurposing in the Public Domain
To be published
|
|
6S1I
 
 | | Crystal Structure of DYRK1A with small molecule inhibitor | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 1A, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Sorrell, F.J, Henderson, S.H, Redondo, C, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Elkins, J.M. | | Deposit date: | 2019-06-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Mining Public Domain Data to Develop Selective DYRK1A Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
6S1H
 
 | | Crystal Structure of DYRK1A with small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, SULFATE ION, ... | | Authors: | Sorrell, F.J, Henderson, S.H, Redondo, C, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Elkins, J.M. | | Deposit date: | 2019-06-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Kinase Scaffold Repurposing in the Public Domain
To be published
|
|
6S1J
 
 | | Crystal Structure of DYRK1A with small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 3-[2-[(3~{S})-3-fluoranylpyrrolidin-1-yl]pyrimidin-4-yl]pyrazolo[1,5-b]pyridazine, DIMETHYL SULFOXIDE, ... | | Authors: | Sorrell, F.J, Henderson, S.H, Redondo, C, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Elkins, J.M. | | Deposit date: | 2019-06-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.408 Å) | | Cite: | Kinase Scaffold Repurposing in the Public Domain
To be published
|
|
6S11
 
 | | Crystal Structure of DYRK1A with small molecule inhibitor | | Descriptor: | 6-pyridin-4-yl-3-[3-(trifluoromethyloxy)phenyl]imidazo[1,2-b]pyridazine, CHLORIDE ION, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Sorrell, F.J, Henderson, S.H, Redondo, C, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Elkins, J.M. | | Deposit date: | 2019-06-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.445 Å) | | Cite: | Kinase Scaffold Repurposing in the Public Domain
To be published
|
|
6S14
 
 | | Crystal Structure of DYRK1A with small molecule inhibitor | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 1A, SULFATE ION, ~{N}-cyclopropyl-~{N}-methyl-4-pyrazolo[1,5-b]pyridazin-3-yl-pyrimidin-2-amine | | Authors: | Sorrell, F.J, Henderson, S.H, Redondo, C, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Elkins, J.M. | | Deposit date: | 2019-06-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Kinase Scaffold Repurposing in the Public Domain
To be published
|
|
6S17
 
 | | Crystal Structure of DYRK1A with small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, SULFATE ION, ... | | Authors: | Sorrell, F.J, Henderson, S.H, Redondo, C, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Elkins, J.M. | | Deposit date: | 2019-06-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Kinase Scaffold Repurposing in the Public Domain
To be published
|
|
4X5S
 
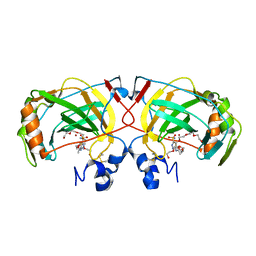 | | The crystal structure of an alpha carbonic anhydrase from the extremophilic bacterium Sulfurihydrogenibium azorense. | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase (Carbonate dehydratase), ... | | Authors: | De Simone, G, Alterio, V, Di Fiore, A. | | Deposit date: | 2014-12-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the most catalytically effective carbonic anhydrase enzyme known, SazCA from the thermophilic bacterium Sulfurihydrogenibium azorense.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
5MY8
 
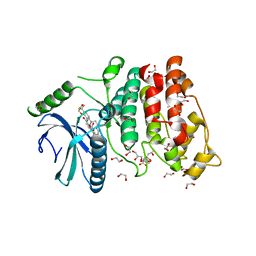 | | Crystal structure of SRPK1 in complex with SPHINX31 | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-01-25 | | Release date: | 2017-05-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of Potent, Selective SRPK1 Inhibitors as Potential Topical Therapeutics for Neovascular Eye Disease.
ACS Chem. Biol., 12, 2017
|
|
5MYV
 
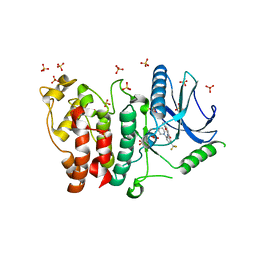 | | Crystal structure of SRPK2 in complex with compound 1 | | Descriptor: | 5-methyl-~{N}-[2-(4-methylpiperazin-1-yl)-5-(trifluoromethyl)phenyl]furan-2-carboxamide, DIMETHYL SULFOXIDE, SRSF protein kinase 2,SRSF protein kinase 2, ... | | Authors: | Chaikuad, A, Pike, A.C.W, Savitsky, P, von Delft, F, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-01-29 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Development of Potent, Selective SRPK1 Inhibitors as Potential Topical Therapeutics for Neovascular Eye Disease.
ACS Chem. Biol., 12, 2017
|
|
3NE0
 
 | |
5IX9
 
 | | Cell surface anchoring domain | | Descriptor: | Antifreeze protein | | Authors: | Guo, S, Langelaan, D. | | Deposit date: | 2016-03-23 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice.
Sci Adv, 3, 2017
|
|
5J6Y
 
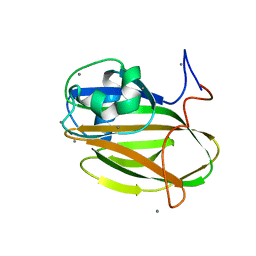 | | Crystal structure of PA14 domain of MpAFP Antifreeze protein | | Descriptor: | Antifreeze protein, CALCIUM ION, alpha-D-glucopyranose, ... | | Authors: | Guo, S. | | Deposit date: | 2016-04-05 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice.
Sci Adv, 3, 2017
|
|
5JUH
 
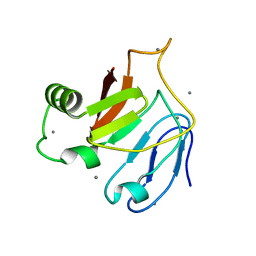 | | Crystal structure of C-terminal domain (RV) of MpAFP | | Descriptor: | Antifreeze protein, CALCIUM ION | | Authors: | Guo, S. | | Deposit date: | 2016-05-10 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice.
Sci Adv, 3, 2017
|
|
5K8G
 
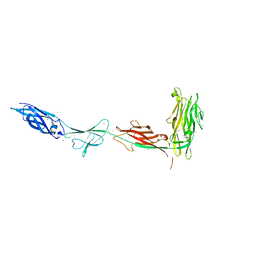 | |
5IRB
 
 | | Structural insight into host cell surface retention of a 1.5-MDa bacterial ice-binding adhesin | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Guo, S, Phippen, S, Campbell, R, Davies, P. | | Deposit date: | 2016-03-12 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice.
Sci Adv, 3, 2017
|
|
