2QV8
 
 | |
2RET
 
 | | The crystal structure of a binary complex of two pseudopilins: EpsI and EpsJ from the Type 2 Secretion System of Vibrio vulnificus | | Descriptor: | CHLORIDE ION, EpsJ, Pseudopilin EpsI, ... | | Authors: | Yanez, M.E, Korotkov, K.V, Abendroth, J, Hol, W.G.J. | | Deposit date: | 2007-09-27 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The crystal structure of a binary complex of two pseudopilins: EpsI and EpsJ from the type 2 secretion system of Vibrio vulnificus.
J.Mol.Biol., 375, 2008
|
|
8DOF
 
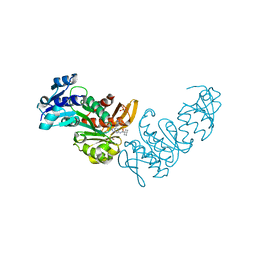 | | Pseudomonas aeruginosa MurC with WYH9-2-P - OSA_001044 | | Descriptor: | (2R)-2-cyclohexyl-2-[(4-{[5-(propan-2-yl)-1H-pyrazol-3-yl]amino}-1H-pyrazolo[3,4-d]pyrimidin-6-yl)amino]ethan-1-ol, 1,2-ETHANEDIOL, SULFATE ION, ... | | Authors: | Horanyi, P.S, Wang, Y, Todd, M.H, Abendroth, J, Edwards, T, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-07-13 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Pseudomonas aeruginosa MurC with WYH9-2-P - OSA_001044
To Be Published
|
|
5E0I
 
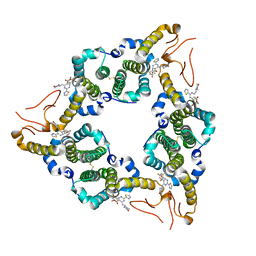 | |
5DLC
 
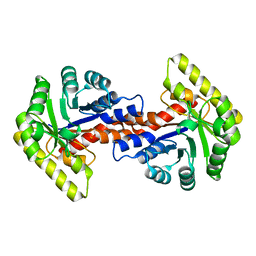 | |
4OGY
 
 | | Crystal structure of Fab DX-2930 in complex with human plasma kallikrein at 2.1 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, DX-2930 HEAVY CHAIN, DX-2930 LIGHT CHAIN, ... | | Authors: | Edwards, T.E, Clifton, M.C, Abendroth, J, Nixon, A, Ladner, R. | | Deposit date: | 2014-01-16 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of plasma kallikrein by a highly specific active site blocking antibody.
J.Biol.Chem., 289, 2014
|
|
4OGX
 
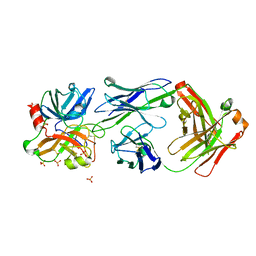 | | Crystal structure of Fab DX-2930 in complex with human plasma kallikrein at 2.4 Angstrom resolution | | Descriptor: | DX-2930 HEAVY CHAIN, DX-2930 LIGHT CHAIN, Plasma kallikrein, ... | | Authors: | Edwards, T.E, Clifton, M.C, Abendroth, J, Nixon, A, Ladner, R. | | Deposit date: | 2014-01-16 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition of plasma kallikrein by a highly specific active site blocking antibody.
J.Biol.Chem., 289, 2014
|
|
4KMS
 
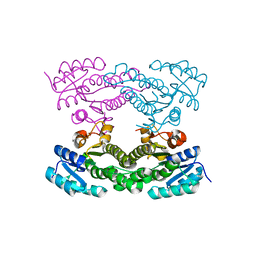 | | Crystal structure of Acetoacetyl-CoA reductase from Rickettsia felis | | Descriptor: | Acetoacetyl-CoA reductase | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID), Abendroth, J, Lukacs, C, Edwards, T.E, Lorimer, D. | | Deposit date: | 2013-05-08 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of acetoacetyl-CoA reductase from Rickettsia felis.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
4NI5
 
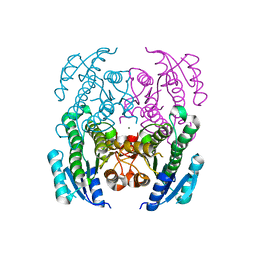 | | Crystal Structure of a Short Chain Dehydrogenase from Brucella suis | | Descriptor: | MAGNESIUM ION, Oxidoreductase, short-chain dehydrogenase/reductase family protein | | Authors: | Dranow, D.M, Abendroth, J, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2013-11-05 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of a Short Chain Dehydrogenase from Brucella suis
TO BE PUBLISHED
|
|
4XIN
 
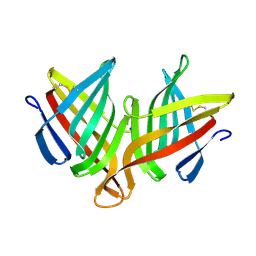 | |
4XUX
 
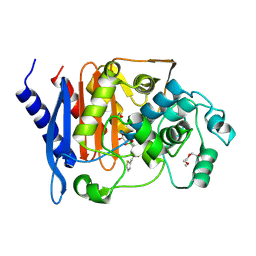 | | Structure of ampC bound to RPX-7009 at 1.75 A | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, Beta-lactamase, ... | | Authors: | Clifton, M.C, Abendroth, J. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of a Cyclic Boronic Acid beta-Lactamase Inhibitor (RPX7009) with Utility vs Class A Serine Carbapenemases.
J.Med.Chem., 58, 2015
|
|
3UWB
 
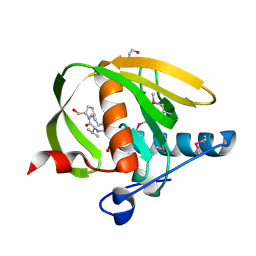 | | Crystal structure of a probable peptide deformylase from strucynechococcus phage S-SSM7 in complex with actinonin | | Descriptor: | 1,2-ETHANEDIOL, ACTINONIN, CHLORIDE ION, ... | | Authors: | Lorimer, D, Abendroth, J, Edwards, T.E, Burgin, A, Segall, A, Rohwer, F. | | Deposit date: | 2011-12-01 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and function of a cyanophage-encoded peptide deformylase.
ISME J, 7, 2013
|
|
3UWA
 
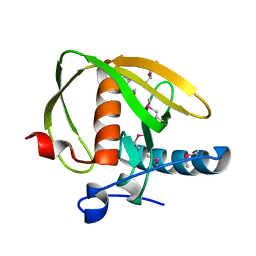 | | Crystal structure of a probable peptide deformylase from synechococcus phage S-SSM7 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, RIIA-RIIB membrane-associated protein, ZINC ION | | Authors: | Lorimer, D, Abendroth, J, Edwards, T.E, Burgin, A, Segall, A, Rohwer, F. | | Deposit date: | 2011-12-01 | | Release date: | 2013-01-09 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and function of a cyanophage-encoded peptide deformylase.
ISME J, 7, 2013
|
|
7UP6
 
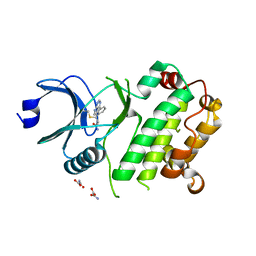 | | Crystal structure of C-terminal domain of MSK1 in complex with in covalently bound literature RSK2 inhibitor pyrrolopyrimidine cyanoacrylamide compound 25 (co-crystal) | | Descriptor: | (E)-3-(3-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)phenyl)-2-cyanoacrylamide bound form, OXAMIC ACID, Ribosomal protein S6 kinase alpha-5 | | Authors: | Yano, J.K, Abendroth, J, Hall, A. | | Deposit date: | 2022-04-14 | | Release date: | 2022-08-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery and Characterization of a Novel Series of Chloropyrimidines as Covalent Inhibitors of the Kinase MSK1.
Acs Med.Chem.Lett., 13, 2022
|
|
7UP4
 
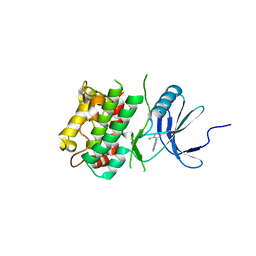 | | Crystal structure of C-terminal Domain of MSK1 in complex with covalently bound pyrrolopyrimidine compound 20 (co-crystal) | | Descriptor: | (5M)-5-(2,5-dichloropyrimidin-4-yl)-5H-pyrrolo[3,2-d]pyrimidine, Ribosomal protein S6 kinase alpha-5 | | Authors: | Yano, J.K, Abendroth, J, Hall, A. | | Deposit date: | 2022-04-14 | | Release date: | 2022-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery and Characterization of a Novel Series of Chloropyrimidines as Covalent Inhibitors of the Kinase MSK1.
Acs Med.Chem.Lett., 13, 2022
|
|
7UP8
 
 | |
7UP7
 
 | | Crystal structure of C-terminal Domain of MSK1 in complex with covalently bound with literature RSK2 inhibitor indazole cyanoacrylamide compound 26 (soak) | | Descriptor: | (2S)-2-cyano-N-(1-hydroxy-2-methylpropan-2-yl)-3-[3-(3,4,5-trimethoxyphenyl)-1H-indazol-5-yl]propanamide, Ribosomal protein S6 kinase alpha-5 | | Authors: | Yano, J.K, Abendroth, J, Hall, A. | | Deposit date: | 2022-04-14 | | Release date: | 2022-07-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Characterization of a Novel Series of Chloropyrimidines as Covalent Inhibitors of the Kinase MSK1.
Acs Med.Chem.Lett., 13, 2022
|
|
6U94
 
 | | Structure of RND efflux system, outer membrane lipoprotein, NodT family from Burkholderia mallei ATCC 23344 | | Descriptor: | GLYCEROL, RND efflux system, outer membrane lipoprotein, ... | | Authors: | Horanyi, P.S, Fox III, D, Abendroth, J, Lorimer, D, Edwards, T, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-09-06 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of RND efflux system, outer membrane lipoprotein, NodT family from Burkholderia mallei ATCC 23344
To Be Published
|
|
4PBC
 
 | |
4EYE
 
 | |
4PN3
 
 | | Crystal structure of 3-hydroxyacyl-CoA-dehydrogenase from Brucella melitensis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyacyl-CoA dehydrogenase | | Authors: | Lukacs, C.M, Abendroth, J, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2014-05-22 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of 3-hydroxyacyl-CoA-dehydrogenase from Brucella melitensis
To Be Published
|
|
6CHD
 
 | |
4PFZ
 
 | |
3U0B
 
 | | Crystal structure of an oxidoreductase from Mycobacterium smegmatis | | Descriptor: | Oxidoreductase, short chain dehydrogenase/reductase family protein, SODIUM ION | | Authors: | Arakaki, T.L, Staker, B.L, Clifton, M.C, Abendroth, J, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2011-09-28 | | Release date: | 2011-10-05 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional characterization of FabG4 from Mycolicibacterium smegmatis.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
4TMD
 
 | | X-ray structure of Putative uncharacterized protein (Rv0999 ortholog) from Mycobacterium smegmatis | | Descriptor: | IODIDE ION, Uncharacterized protein | | Authors: | Horanyi, P.S, Dranow, D.M, Abendroth, J, Lorimer, D, Edwards, T, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2014-06-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of Putative uncharacterized protein (Rv0999 ortholog) from Mycobacterium smegmatis
To Be Published
|
|
