7E2G
 
 | | Cryo-EM structure of hDisp1NNN-3C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Protein dispatched homolog 1,Protein dispatched homolog 1 | | Authors: | Li, W, Wang, L, Gong, X. | | Deposit date: | 2021-02-05 | | Release date: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Structural insights into proteolytic activation of the human Dispatched1 transporter for Hedgehog morphogen release.
Nat Commun, 12, 2021
|
|
7E2I
 
 | | Cryo-EM structure of hDisp1NNN-ShhN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Protein dispatched homolog 1, ... | | Authors: | Li, W, Wang, L, Gong, X. | | Deposit date: | 2021-02-05 | | Release date: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Structural insights into proteolytic activation of the human Dispatched1 transporter for Hedgehog morphogen release.
Nat Commun, 12, 2021
|
|
2JAS
 
 | | Structure of deoxyadenosine kinase from M.mycoides with bound dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DEOXYGUANOSINE KINASE, MAGNESIUM ION | | Authors: | Welin, M, Wang, L, Eriksson, S, Eklund, H. | | Deposit date: | 2006-11-30 | | Release date: | 2007-01-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-Function Analysis of a Bacterial Deoxyadenosine Kinase Reveals the Basis for Substrate Specificity.
J.Mol.Biol., 366, 2007
|
|
2J9R
 
 | | Thymidine kinase from B. anthracis in complex with dT. | | Descriptor: | PHOSPHATE ION, THYMIDINE, THYMIDINE KINASE, ... | | Authors: | Kosinska, U, Carnrot, C, Sandrini, M.P.B, Clausen, A.R, Wang, L, Piskur, J, Eriksson, S, Eklund, H. | | Deposit date: | 2006-11-15 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Studies of Thymidine Kinases from Bacillus Anthracis and Bacillus Cereus Provide Insights Into Quaternary Structure and Conformational Changes Upon Substrate Binding
FEBS J., 274, 2007
|
|
2MI2
 
 | |
2JAT
 
 | | Structure of deoxyadenosine kinase from M.mycoides with products dcmp and a flexible dcdp bound | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, DEOXYGUANOSINE KINASE, MAGNESIUM ION, ... | | Authors: | Welin, M, Wang, L, Eriksson, S, Eklund, H. | | Deposit date: | 2006-11-30 | | Release date: | 2007-01-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Function Analysis of a Bacterial Deoxyadenosine Kinase Reveals the Basis for Substrate Specificity.
J.Mol.Biol., 366, 2007
|
|
2JA1
 
 | | Thymidine kinase from B. cereus with TTP bound as phosphate donor. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, THYMIDINE KINASE, THYMIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Kosinska, U, Carnrot, C, Sandrini, M.P.B, Clausen, A.R, Wang, L, Piskur, J, Eriksson, S, Eklund, H. | | Deposit date: | 2006-11-17 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Studies of Thymidine Kinases from Bacillus Anthracis and Bacillus Cereus Provide Insights Into Quaternary Structure and Conformational Changes Upon Substrate Binding
FEBS J., 274, 2007
|
|
2M5K
 
 | | Atomic-resolution structure of a doublet cross-beta amyloid fibril | | Descriptor: | Transthyretin | | Authors: | Fitzpatrick, A.W.P, Debelouchina, G.T, Bayro, M.J, Clare, D.K, Caporini, M.A, Bajaj, V.S, Jaroniec, C.P, Wang, L, Ladizhansky, V, Muller, S, MacPhee, C.E, Waudby, C.A, Mott, H.R, de Simone, A, Knowles, T.P.J, Saibil, H.R, Vendruscolo, M, Orlova, E.V, Griffin, R.G, Dobson, C.M. | | Deposit date: | 2013-02-27 | | Release date: | 2013-12-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (12.7 Å), SOLID-STATE NMR | | Cite: | Atomic structure and hierarchical assembly of a cross-beta amyloid fibril.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2M5M
 
 | | Atomic-resolution structure of a triplet cross-beta amyloid fibril | | Descriptor: | Transthyretin | | Authors: | Fitzpatrick, A.W.P, Debelouchina, G.T, Bayro, M.J, Clare, D.K, Caporini, M.A, Bajaj, V.S, Jaroniec, C.P, Wang, L, Ladizhansky, V, Muller, S, MacPhee, C.E, Waudby, C.A, Mott, H.R, de Simone, A, Knowles, T.P.J, Saibil, H.R, Vendruscolo, M, Orlova, E.V, Griffin, R.G, Dobson, C.M. | | Deposit date: | 2013-02-27 | | Release date: | 2013-12-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (12.2 Å), SOLID-STATE NMR | | Cite: | Atomic structure and hierarchical assembly of a cross-beta amyloid fibril.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2JAQ
 
 | | Structure of deoxyadenosine kinase from M. mycoides with bound dCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DEOXYGUANOSINE KINASE | | Authors: | Welin, M, Wang, L, Eriksson, S, Eklund, H. | | Deposit date: | 2006-11-30 | | Release date: | 2007-01-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Function Analysis of a Bacterial Deoxyadenosine Kinase Reveals the Basis for Substrate Specificity.
J.Mol.Biol., 366, 2007
|
|
2M5N
 
 | | Atomic-resolution structure of a cross-beta protofilament | | Descriptor: | Transthyretin | | Authors: | Fitzpatrick, A.W.P, Debelouchina, G.T, Bayro, M.J, Clare, D.K, Caporini, M.A, Bajaj, V.S, Jaroniec, C.P, Wang, L, Ladizhansky, V, Muller, S, MacPhee, C.E, Waudby, C.A, Mott, H.R, de Simone, A, Knowles, T.P.J, Saibil, H.R, Vendruscolo, M, Orlova, E.V, Griffin, R.G, Dobson, C.M. | | Deposit date: | 2013-02-27 | | Release date: | 2013-07-17 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic structure and hierarchical assembly of a cross-{beta} amyloid fibril.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2OH4
 
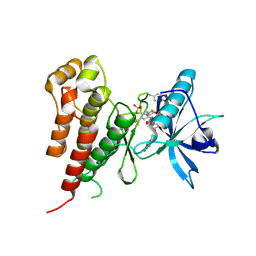 | | Crystal structure of Vegfr2 with a benzimidazole-urea inhibitor | | Descriptor: | METHYL (5-{4-[({[2-FLUORO-5-(TRIFLUOROMETHYL)PHENYL]AMINO}CARBONYL)AMINO]PHENOXY}-1H-BENZIMIDAZOL-2-YL)CARBAMATE, SULFATE ION, Vascular endothelial growth factor receptor 2 | | Authors: | Nolte, R.T, Wang, L. | | Deposit date: | 2007-01-09 | | Release date: | 2007-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of Novel Benzimidazoles as Potent Inhibitors of TIE-2 and VEGFR-2 Tyrosine Kinase Receptors.
J.Med.Chem., 50, 2007
|
|
7VZB
 
 | | Cryo-EM structure of C22:0-CoA bound human very long-chain fatty acid ABC transporter ABCD1 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Peroxisomal Membrane Protein related,ATP-binding cassette sub-family D member 1, S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] docosanethioate | | Authors: | Chen, Z.P, Xu, D, Wang, L, Mao, Y.X, Yang, L, Cheng, M.T, Hou, W.T, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2021-11-15 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structural basis of substrate recognition and translocation by human very long-chain fatty acid transporter ABCD1.
Nat Commun, 13, 2022
|
|
7FAT
 
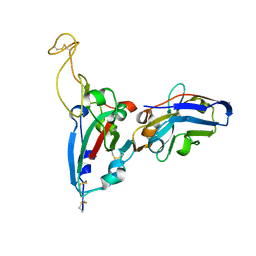 | | Structure Determination of the RBD-NB1A7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, nb_1A7 | | Authors: | Geng, Y, Shi, Z.Z, Li, X.Y, Wang, L, Sun, Z.C, Zhang, H.W, Chen, X.C. | | Deposit date: | 2021-07-07 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis of nanobodies neutralizing SARS-CoV-2 variants
Structure, 30, 2022
|
|
7FAU
 
 | | Structure Determination of the NB1B11-RBD Complex | | Descriptor: | NB_1B11, Spike protein S1, ZINC ION | | Authors: | Shi, Z.Z, Li, X.X, Wang, L, Sun, Z.C, Zhang, H.W, Chen, X.C, Cui, Q.Q, Qiao, H.R, Lan, Z.Y, Zhang, X. | | Deposit date: | 2021-07-07 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis of nanobodies neutralizing SARS-CoV-2 variants.
Structure, 30, 2022
|
|
7XC6
 
 | |
7WWS
 
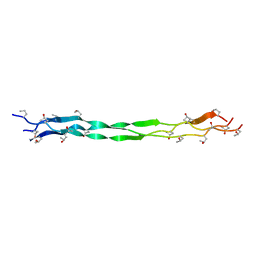 | | Structure of a triple-helix region of human collagen type III from Trautec | | Descriptor: | Collagen alpha-1(III) chain | | Authors: | Qian, S, Li, H, Fan, X, Tian, X, Li, J, Wang, L, Chu, Y. | | Deposit date: | 2022-02-14 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of a triple-helix region of human collagen type III from Trautec
To Be Published
|
|
7WWR
 
 | | Structure of a triple-helix region of human collagen type III from Trautec | | Descriptor: | Collagen alpha-1(III) chain | | Authors: | Qian, S, Li, H, Fan, X, Tian, X, Li, J, Wang, L, Chu, Y. | | Deposit date: | 2022-02-14 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of a triple-helix region of human collagen type III from Trautec
To Be Published
|
|
7XAN
 
 | | Structure of a triple-helix region of human collagen type III from Trautec | | Descriptor: | Collagen alpha-1(III) chain | | Authors: | Qian, S, Li, H, Fan, X, Tian, X, Li, J, Wang, L, Chu, Y. | | Deposit date: | 2022-03-18 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a triple-helix region of human collagen type III from Trautec
To Be Published
|
|
7WVK
 
 | | Crystal structure of human WDR5 in complex with compound 19 | | Descriptor: | 1,2-ETHANEDIOL, 1-[2,5-bis(chloranyl)phenyl]sulfonylbenzimidazole, GLYCEROL, ... | | Authors: | Han, Q.L, Zhang, X.L, Wang, L, Ren, P.X, Cao, Y, Li, K, Bai, F. | | Deposit date: | 2022-02-10 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Discovery, evaluation and mechanism study of WDR5-targeted small molecular inhibitors for neuroblastoma.
Acta Pharmacol.Sin., 44, 2023
|
|
7CYE
 
 | | Cryo-EM structure of sodium-dependent bicarbonate transporter SbtA from Synechocystis sp. PCC 6803 | | Descriptor: | Slr1512 protein | | Authors: | Liu, X.Y, Jiang, Y.L, Wang, L, Hou, W.T, Chen, Y, Zhou, C.Z. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structures of cyanobacterial bicarbonate transporter SbtA and its complex with PII-like SbtB.
Cell Discov, 7, 2021
|
|
7CYF
 
 | | Cryo-EM structure of bicarbonate transporter SbtA in complex with PII-like signaling protein SbtB from Synechocystis sp. PCC 6803 | | Descriptor: | ADENOSINE MONOPHOSPHATE, Membrane-associated protein slr1513, SODIUM ION, ... | | Authors: | Liu, X.Y, Jiang, Y.L, Wang, L, Hou, W.T, Chen, Y, Zhou, C.Z. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-23 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structures of cyanobacterial bicarbonate transporter SbtA and its complex with PII-like SbtB.
Cell Discov, 7, 2021
|
|
7DVQ
 
 | | Cryo-EM Structure of the Activated Human Minor Spliceosome (minor Bact Complex) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'-O-[(S)-hydroxy{[(R)-hydroxy{[(S)-hydroxy(methoxy)phosphoryl]oxy}phosphoryl]oxy}phosphoryl]guanosine, Armadillo repeat-containing protein 7, ... | | Authors: | Bai, R, Wan, R, Wang, L, Xu, K, Zhang, Q, Lei, J, Shi, Y. | | Deposit date: | 2021-01-14 | | Release date: | 2021-03-31 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure of the activated human minor spliceosome.
Science, 371, 2021
|
|
7D1G
 
 | | Crystal structure of Glyceraldehyde-3-Phosphate Dehydrogenase GAPDH from Clostridium beijerinckii | | Descriptor: | BETA-MERCAPTOETHANOL, Glyceraldehyde-3-phosphate dehydrogenase, MAGNESIUM ION | | Authors: | Chen, Y, Lan, J, Liu, W, Wang, L, Xu, Y. | | Deposit date: | 2020-09-14 | | Release date: | 2021-03-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of Glyceraldehyde-3-Phosphate Dehydrogenase GAPDH from Clostridium beijerinckii
To Be Published
|
|
7CQI
 
 | | Cryo-EM structure of the substrate-bound SPT-ORMDL3 complex | | Descriptor: | ORM1-like protein 3, Serine palmitoyltransferase 1, Serine palmitoyltransferase 2, ... | | Authors: | Li, S.S, Xie, T, Wang, L, Gong, X. | | Deposit date: | 2020-08-11 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into the assembly and substrate selectivity of human SPT-ORMDL3 complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
