8HZV
 
 | | The crystal structure of a Radical SAM Enzyme DesII | | Descriptor: | GLYCEROL, IRON/SULFUR CLUSTER, METHIONINE, ... | | Authors: | Hou, X.L, Zhou, J.H. | | Deposit date: | 2023-01-09 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.33001685 Å) | | Cite: | Mechanistic Insights from the Crystal Structure and Computational Analysis of the Radical SAM Deaminase DesII.
Adv Sci, 2024
|
|
7C7X
 
 | |
7CV9
 
 | | RNA methyltransferase METTL4 | | Descriptor: | GLYCEROL, Methyltransferase-like protein 2, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Luo, Q, Ma, J. | | Deposit date: | 2020-08-25 | | Release date: | 2021-09-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.455 Å) | | Cite: | Structural insights into molecular mechanism for N6-adenosine methylation by MT-A70 family methyltransferase METTL4
Nat Commun, 13, 2022
|
|
7CVA
 
 | | RNA methyltransferase METTL4 | | Descriptor: | Methyltransferase-like protein 2 | | Authors: | Luo, Q, Ma, J. | | Deposit date: | 2020-08-25 | | Release date: | 2021-09-01 | | Last modified: | 2022-10-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into molecular mechanism for N6-adenosine methylation by MT-A70 family methyltransferase METTL4
Nat Commun, 13, 2022
|
|
7CV7
 
 | | RNA methyltransferase METTL4 | | Descriptor: | Methyltransferase-like protein 2, S-ADENOSYLMETHIONINE | | Authors: | Luo, Q, Ma, J. | | Deposit date: | 2020-08-25 | | Release date: | 2021-09-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into molecular mechanism for N6-adenosine methylation by MT-A70 family methyltransferase METTL4
Nat Commun, 13, 2022
|
|
7CV6
 
 | | RNA methyltransferase METTL4 | | Descriptor: | 2'-O-methyladenosine 5'-(dihydrogen phosphate), Methyltransferase-like protein 2, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Luo, Q, Ma, J. | | Deposit date: | 2020-08-25 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural insights into molecular mechanism for N6-adenosine methylation by MT-A70 family methyltransferase METTL4
Nat Commun, 13, 2022
|
|
7CV8
 
 | | RNA methyltransferase METTL4 | | Descriptor: | GLYCEROL, Methyltransferase-like protein 2, SINEFUNGIN | | Authors: | Luo, Q, Ma, J. | | Deposit date: | 2020-08-25 | | Release date: | 2021-09-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural insights into molecular mechanism for N6-adenosine methylation by MT-A70 family methyltransferase METTL4
Nat Commun, 13, 2022
|
|
8FDL
 
 | | Human Hemoglobin with Nitrosochloramphenicol | | Descriptor: | GLYCEROL, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Powell, S.M, Richter-Addo, G.B, Thomas, L.M. | | Deposit date: | 2022-12-03 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structural investigations of heme protein derivatives resulting from reactions of aryl- and alkylhydroxylamines with human hemoglobin.
J.Inorg.Biochem., 246, 2023
|
|
8FDN
 
 | | Human Hemoglobin with N-tertbutylhydroxylamine | | Descriptor: | GLYCEROL, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Powell, S.M, Richter-Addo, G.B, Thomas, L.M. | | Deposit date: | 2022-12-03 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structural investigations of heme protein derivatives resulting from reactions of aryl- and alkylhydroxylamines with human hemoglobin.
J.Inorg.Biochem., 246, 2023
|
|
8FDM
 
 | | Human Hemoglobin in Complex with Nitrosomethane | | Descriptor: | GLYCEROL, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Powell, S.M, Richter-Addo, G.B, Thomas, L.M. | | Deposit date: | 2022-12-03 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structural investigations of heme protein derivatives resulting from reactions of aryl- and alkylhydroxylamines with human hemoglobin.
J.Inorg.Biochem., 246, 2023
|
|
7BP2
 
 | |
7BP5
 
 | |
7F18
 
 | | Crystal Structure of a mutant of acid phosphatase from Pseudomonas aeruginosa (Q57H/W58P/D135R) | | Descriptor: | Acid phosphatase | | Authors: | Xu, X, Hou, X.D, Song, W, Yin, D.J, Rao, Y.J, Liu, L.M. | | Deposit date: | 2021-06-08 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Local Electric Field Modulated Reactivity of Pseudomonas aeruginosa Acid Phosphatase for Enhancing Phosphorylation of l-Ascorbic Acid
Acs Catalysis, 11, 2021
|
|
7F17
 
 | | Crystal Structure of acid phosphatase | | Descriptor: | Acid phosphatase | | Authors: | Xu, X, Hou, X.D, Song, W, Rao, Y.J, Liu, L.M, Wu, J. | | Deposit date: | 2021-06-08 | | Release date: | 2021-10-27 | | Last modified: | 2022-05-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Local Electric Field Modulated Reactivity of Pseudomonas aeruginosa Acid Phosphatase for Enhancing Phosphorylation of l-Ascorbic Acid
Acs Catalysis, 11, 2021
|
|
3NNL
 
 | | Halogenase domain from CurA module (crystal form III) | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, CurA, ... | | Authors: | Khare, D, Smith, J.L. | | Deposit date: | 2010-06-23 | | Release date: | 2010-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.883 Å) | | Cite: | Conformational switch triggered by alpha-ketoglutarate in a halogenase of curacin A biosynthesis
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3NNF
 
 | | Halogenase domain from CurA module with Fe, chloride, and alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, CurA, ... | | Authors: | Khare, D, Smith, J.L. | | Deposit date: | 2010-06-23 | | Release date: | 2010-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Conformational switch triggered by alpha-ketoglutarate in a halogenase of curacin A biosynthesis
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7URF
 
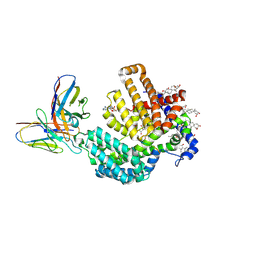 | | Human HHAT H379C in complex with SHH N-terminal peptide | | Descriptor: | 3H02 heavy chain, 3H02 light chain, Digitonin, ... | | Authors: | Liu, Y, Qi, X, Li, X. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanisms and inhibition of Porcupine-mediated Wnt acylation.
Nature, 607, 2022
|
|
7URE
 
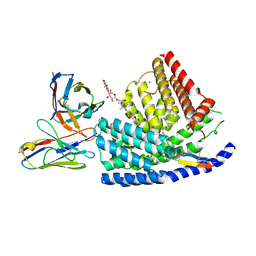 | | Human PORCN in complex with palmitoleoylated WNT3A peptide | | Descriptor: | 2C11 heavy chain, 2C11 light chain, Digitonin, ... | | Authors: | Liu, Y, Qi, X, Li, X. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Mechanisms and inhibition of Porcupine-mediated Wnt acylation.
Nature, 607, 2022
|
|
7URC
 
 | | Human PORCN in complex with LGK974 | | Descriptor: | 2-[(2P)-2',3-dimethyl[2,4'-bipyridin]-5-yl]-N-[(5P)-5-(pyrazin-2-yl)pyridin-2-yl]acetamide, 2C11 heavy chain, 2C11 light chain, ... | | Authors: | Liu, Y, Qi, X, Li, X. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Mechanisms and inhibition of Porcupine-mediated Wnt acylation.
Nature, 607, 2022
|
|
7URA
 
 | | Human PORCN in complex with Palmitoleoyl-CoA | | Descriptor: | 2C11 heavy chain, 2C11 light chain, CHOLESTEROL, ... | | Authors: | Liu, Y, Qi, X, Li, X. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Mechanisms and inhibition of Porcupine-mediated Wnt acylation.
Nature, 607, 2022
|
|
7URD
 
 | | Human PORCN in complex with LGK974 and WNT3A peptide | | Descriptor: | 2-[(2P)-2',3-dimethyl[2,4'-bipyridin]-5-yl]-N-[(5P)-5-(pyrazin-2-yl)pyridin-2-yl]acetamide, 2C11 heavy chain, 2C11 light chain, ... | | Authors: | Liu, Y, Qi, X, Li, X. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Mechanisms and inhibition of Porcupine-mediated Wnt acylation.
Nature, 607, 2022
|
|
3NNM
 
 | |
7TDQ
 
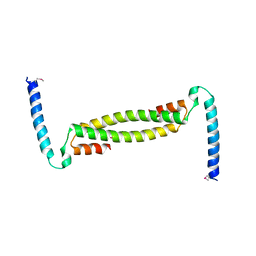 | |
7TOL
 
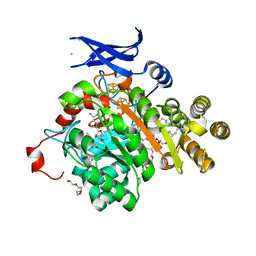 | | X-ray crystal structure of glycerol dibiphytanyl glycerol tetraether - macrocyclic archaeol synthase (GDGT-MAS) from Methanocaldococcus jannaschii with archaeal lipid, 5'deoxyadenosine, and methionine bound | | Descriptor: | 3-PHOSPHORYL-[1,2-DI-PHYTANYL]GLYCEROL, 3-[GLYCEROLYLPHOSPHONYL]-[1,2-DI-PHYTANYL]GLYCEROL, 5'-DEOXYADENOSINE, ... | | Authors: | Lloyd, C.T, Booker, S.J, Boal, A.K. | | Deposit date: | 2022-01-24 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery, structure and mechanism of a tetraether lipid synthase.
Nature, 609, 2022
|
|
7VOC
 
 | | The crystal structure of a Radical SAM Enzyme BlsE involved in the Biosynthesis of Blasticidin S | | Descriptor: | (2~{S},3~{S},4~{S},5~{R},6~{R})-6-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4,5-tris(oxidanyl)oxane-2-carboxylic acid, Cytosylglucuronate decarboxylase, GLYCEROL, ... | | Authors: | Hou, X.L, Zhou, J.H. | | Deposit date: | 2021-10-13 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.62005424 Å) | | Cite: | Radical S -Adenosyl Methionine Enzyme BlsE Catalyzes a Radical-Mediated 1,2-Diol Dehydration during the Biosynthesis of Blasticidin S.
J.Am.Chem.Soc., 144, 2022
|
|
