4LCT
 
 | | Crystal Structure and Versatile Functional Roles of the COP9 Signalosome Subunit 1 | | Descriptor: | COP9 signalosome complex subunit 1, SULFATE ION | | Authors: | Lee, J.-H, Yi, L, Li, J, Schweitzer, K, Borgmann, M, Naumann, M, Wu, H. | | Deposit date: | 2013-06-23 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and versatile functional roles of the COP9 signalosome subunit 1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4LD6
 
 | | PWWP domain of human PWWP Domain-Containing Protein 2B | | Descriptor: | GLYCEROL, PWWP domain-containing protein 2B, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Dombrovski, L, Tempel, W, Loppnau, P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-06-24 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of PWWP domain of human PWWP Domain-Containing Protein 2B
To be Published
|
|
4L5S
 
 | | p202 HIN1 in complex with 12-mer dsDNA | | Descriptor: | 12-mer DNA, Interferon-activable protein 202, SULFATE ION | | Authors: | Yin, Q, Tian, Y, Wu, H. | | Deposit date: | 2013-06-11 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Molecular Mechanism for p202-Mediated Specific Inhibition of AIM2 Inflammasome Activation.
Cell Rep, 4, 2013
|
|
5W1R
 
 | | Cryo-EM structure of DNAPKcs | | Descriptor: | DNA-dependent protein kinase catalytic subunit | | Authors: | Sharif, H, Li, Y, Wu, H. | | Deposit date: | 2017-06-04 | | Release date: | 2017-07-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structure of the DNA-PK holoenzyme.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4LWD
 
 | | Human CARMA1 CARD domain | | Descriptor: | Caspase recruitment domain-containing protein 11, MAGNESIUM ION, SULFATE ION | | Authors: | Zheng, C, Wu, H. | | Deposit date: | 2013-07-26 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Structural Architecture of the CARMA1/Bcl10/MALT1 Signalosome: Nucleation-Induced Filamentous Assembly.
Mol.Cell, 51, 2013
|
|
5Z2S
 
 | | Crystal structure of DUX4-HD2 domain | | Descriptor: | Double homeobox protein 4 | | Authors: | Dong, X, Zhang, W, Wu, H, Huang, J, Zhang, M, Wang, P, Zhang, H, Chen, Z, Chen, S, Meng, G. | | Deposit date: | 2018-01-03 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of DUX4/IGH-driven transactivation.
Leukemia, 32, 2018
|
|
2GTR
 
 | | Human chromodomain Y-like protein | | Descriptor: | Chromodomain Y-like protein | | Authors: | Min, J, Wu, H, Antoshenko, T, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-04-28 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of Human chromodomain Y-like Protein.
To be Published
|
|
2LGG
 
 | | Structure of PHD domain of UHRF1 in complex with H3 peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION, histone H3 peptide | | Authors: | Wang, C, Shen, J, Yang, Z, Chen, P, Zhao, B, Hu, W, Lan, W, Tong, X, Wu, H, Li, G, Cao, C. | | Deposit date: | 2011-07-26 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger.
Cell Res., 21, 2011
|
|
4N4W
 
 | | Structure of the human smoothened receptor in complex with SANT-1. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (E)-N-(4-benzylpiperazin-1-yl)-1-(3,5-dimethyl-1-phenyl-1H-pyrazol-4-yl)methanimine, Cytochrome b(562),Smoothened homolog, ... | | Authors: | Wang, C, Wu, H, Han, G.W, Cherezov, V, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2013-10-08 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for Smoothened receptor modulation and chemoresistance to anticancer drugs.
Nat Commun, 5, 2014
|
|
5YUF
 
 | | Crystal Structure of PML RING tetramer | | Descriptor: | Protein PML, ZINC ION | | Authors: | Wang, P, Benhend, S, Wu, H, Breitenbach, V, Zhen, T, Jollivet, F, Peres, L, Li, Y, Chen, S, Chen, Z, de THE, H, Meng, G. | | Deposit date: | 2017-11-22 | | Release date: | 2018-04-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | RING tetramerization is required for nuclear body biogenesis and PML sumoylation.
Nat Commun, 9, 2018
|
|
4L5T
 
 | | Crystal structure of the tetrameric p202 HIN2 | | Descriptor: | Interferon-activable protein 202 | | Authors: | Yin, Q, Tian, Y, Wu, H. | | Deposit date: | 2013-06-11 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.405 Å) | | Cite: | Molecular Mechanism for p202-Mediated Specific Inhibition of AIM2 Inflammasome Activation.
Cell Rep, 4, 2013
|
|
5WCG
 
 | | SET and MYND Domain Containing protein 2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIMETHYL SULFOXIDE, N-lysine methyltransferase SMYD2, ... | | Authors: | Dong, A, Zeng, H, Walker, J.R, Hutch, A, Seitova, A, Tatlock, J, Kumpf, R, Owen, A, Taylor, A, Casimiro-Garcia, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-30 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The crystal structure of SMYD2 in complex with compound MTF003
to be published
|
|
4O7Q
 
 | |
2K1Z
 
 | |
2LIT
 
 | | NMR Solution Structure of Yeast Iso-1-cytochrome c Mutant P71H in reduced states | | Descriptor: | Cytochrome c iso-1, HEME C | | Authors: | Lan, W, Wang, Z, Yang, Z, Zhu, J, Ying, T, Jiang, X, Zhang, X, Wu, H, Liu, M, Tan, X, Cao, C, Huang, Z.X. | | Deposit date: | 2011-08-31 | | Release date: | 2011-12-07 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Conformational toggling of yeast iso-1-cytochrome C in the oxidized and reduced States.
Plos One, 6, 2011
|
|
4N0U
 
 | | Ternary complex between Neonatal Fc receptor, serum albumin and Fc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Ig gamma-1 chain C region, ... | | Authors: | Oganesyan, V, Wu, H, Dall'Acqua, W.F. | | Deposit date: | 2013-10-02 | | Release date: | 2014-02-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural Insights into Neonatal Fc Receptor-based Recycling Mechanisms.
J.Biol.Chem., 289, 2014
|
|
2LGK
 
 | | NMR Structure of UHRF1 PHD domains in a complex with histone H3 peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION, histone H3 peptide | | Authors: | Wang, C, Shen, J, Yang, Z, Chen, P, Zhao, B, Hu, W, Lan, W, Tong, X, Wu, H, Li, G, Cao, C. | | Deposit date: | 2011-07-28 | | Release date: | 2011-09-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger.
Cell Res., 21, 2011
|
|
5Z2T
 
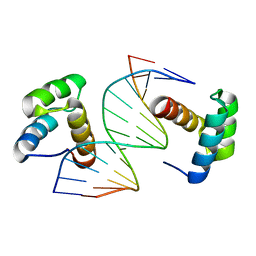 | | Crystal structure of DNA-bound DUX4-HD2 | | Descriptor: | 5'-D(*TP*TP*CP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*TP*T)-3', 5'-D(P*AP*AP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*GP*T)-3', Double homeobox protein 4 | | Authors: | Dong, X, Zhang, W, Wu, H, Huang, J, Zhang, M, Wang, P, Zhang, H, Chen, Z, Chen, S, Meng, G. | | Deposit date: | 2018-01-04 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.623 Å) | | Cite: | Structural basis of DUX4/IGH-driven transactivation.
Leukemia, 32, 2018
|
|
2LIR
 
 | | NMR Solution Structure of Yeast Iso-1-cytochrome c Mutant P71H in oxidized states | | Descriptor: | Cytochrome c iso-1, HEME C | | Authors: | Lan, W, Wang, Z, Yang, Z, Zhu, J, Ying, T, Jiang, X, Zhang, X, Wu, H, Liu, M, Tan, X, Cao, C, Huang, Z.X. | | Deposit date: | 2011-08-31 | | Release date: | 2011-12-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Conformational toggling of yeast iso-1-cytochrome C in the oxidized and reduced States.
Plos One, 6, 2011
|
|
2LGL
 
 | | NMR structure of the UHRF1 PHD domain | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION | | Authors: | Wang, C, Shen, J, Yang, Z, Chen, P, Zhao, B, Hu, W, Lan, W, Tong, X, Wu, H, Li, G, Cao, C. | | Deposit date: | 2011-07-28 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger.
Cell Res., 21, 2011
|
|
2LV9
 
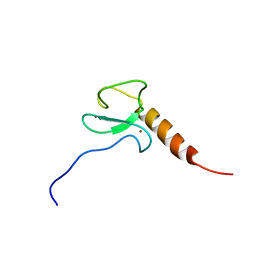 | | Solution NMR structure of the PHD domain of human MLL5, Northeast structural genomics consortium target HR6512A | | Descriptor: | Histone-lysine N-methyltransferase MLL5, ZINC ION | | Authors: | Lemak, A, Yee, A, Houliston, S, Garcia, M, Wu, H, Min, J, Montelione, G.T, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG), Structural Genomics Consortium (SGC), Chaperone-Enabled Studies of Epigenetic Regulation Enzymes (CEBS) | | Deposit date: | 2012-06-29 | | Release date: | 2012-09-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the human MLL5 PHD domain (CASP Target)
To be Published
|
|
2K20
 
 | |
2MB9
 
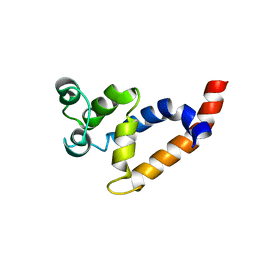 | | Human Bcl10 CARD | | Descriptor: | B-cell lymphoma/leukemia 10 | | Authors: | Zheng, C, Bracken, C, Wu, H. | | Deposit date: | 2013-07-26 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Architecture of the CARMA1/Bcl10/MALT1 Signalosome: Nucleation-Induced Filamentous Assembly.
Mol.Cell, 51, 2013
|
|
2KBK
 
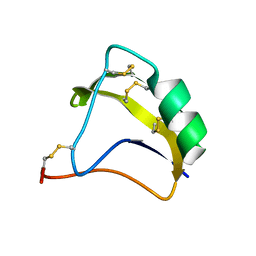 | | Solution Structure of BmK-M10 | | Descriptor: | Neurotoxin BmK-M10 | | Authors: | Zhu, J, Wu, H. | | Deposit date: | 2008-11-28 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of BmK-M10
To be Published
|
|
2M21
 
 | | Solution structure of the Tetrahymena telomerase RNA stem IV terminal loop | | Descriptor: | 5'-R(*GP*GP*CP*GP*AP*UP*AP*CP*AP*CP*UP*AP*UP*UP*UP*AP*UP*CP*GP*CP*C)-3' | | Authors: | Cash, D.D, Richards, R.J, Wu, H, Trantirek, L, O'Connor, C.M, Feigon, J, Collins, K. | | Deposit date: | 2012-12-11 | | Release date: | 2013-03-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural study of elements of Tetrahymena telomerase RNA stem-loop IV domain important for function.
Rna, 12, 2006
|
|
