3O10
 
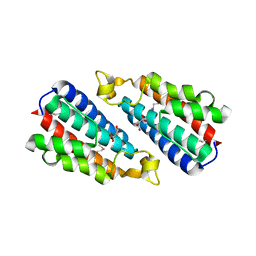 | | Crystal structure of the HEPN domain from human sacsin | | Descriptor: | MALONATE ION, Sacsin | | Authors: | Kozlov, G, Gehring, K. | | Deposit date: | 2010-07-20 | | Release date: | 2011-03-30 | | Last modified: | 2014-07-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of defects in the sacsin HEPN domain responsible for autosomal recessive spastic ataxia of Charlevoix-Saguenay (ARSACS).
J.Biol.Chem., 286, 2011
|
|
6CO4
 
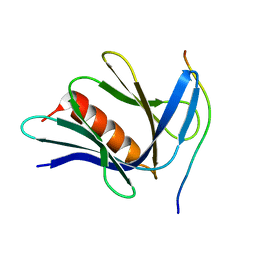 | |
3QK2
 
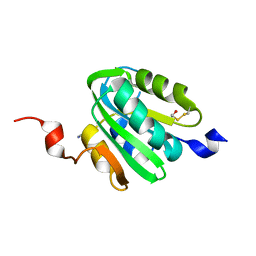 | |
3QN2
 
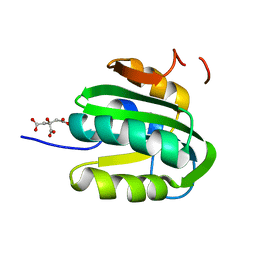 | |
2MBH
 
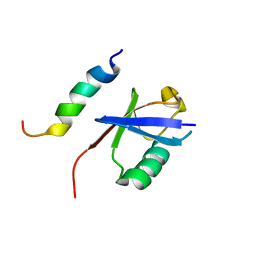 | | NMR structure of EKLF(22-40)/Ubiquitin Complex | | Descriptor: | Krueppel-like factor 1, Ubiquitin | | Authors: | Raiola, L, Omichinski, J.G. | | Deposit date: | 2013-07-31 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of a Noncovalent Complex between Ubiquitin and the Transactivation Domain of the Erythroid-Specific Factor EKLF.
Structure, 21, 2013
|
|
2M64
 
 | | 1H, 13C and 15N Chemical Shift Assignments for Phl p 5a | | Descriptor: | Phlp5 | | Authors: | Goebl, C, Focke, M, Schrank, E, Madl, T, Kosol, S, Madritsch, C, Flicker, S, Valenta, R, Zangger, K, Tjandra, N. | | Deposit date: | 2013-03-21 | | Release date: | 2014-03-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Flexible IgE epitope-containing domains of Phl p 5 cause high allergenic activity.
J. Allergy Clin. Immunol., 140, 2017
|
|
1R6N
 
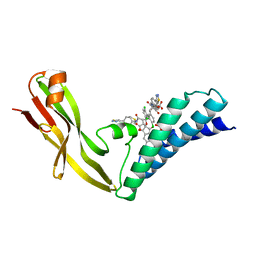 | | HPV11 E2 TAD complex crystal structure | | Descriptor: | 2-METHYL-PROPIONIC ACID, DIMETHYL SULFOXIDE, HPV11 REGULATORY PROTEIN E2, ... | | Authors: | Wang, Y, Coulombe, R. | | Deposit date: | 2003-10-15 | | Release date: | 2004-02-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the E2 Transactivation Domain of Human
Papillomavirus Type 11 Bound to a Protein Interaction Inhibitor
J.Biol.Chem., 279, 2004
|
|
1R6K
 
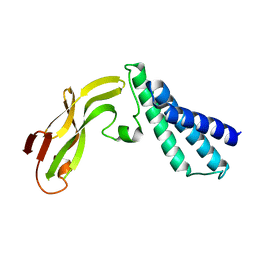 | | HPV11 E2 TAD crystal structure | | Descriptor: | HPV11 REGULATORY PROTEIN E2 | | Authors: | Wang, Y, Coulombe, R. | | Deposit date: | 2003-10-15 | | Release date: | 2004-02-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the E2 Transactivation Domain of Human Papillomavirus Type 11 Bound to a Protein Interaction Inhibitor
J.Biol.Chem., 279, 2004
|
|
1SKF
 
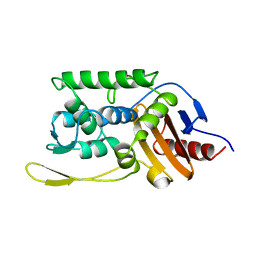 | | CRYSTAL STRUCTURE OF THE STREPTOMYCES K15 DD-TRANSPEPTIDASE | | Descriptor: | D-ALANYL-D-ALANINE TRANSPEPTIDASE | | Authors: | Fonze, E, Charlier, P. | | Deposit date: | 1998-08-20 | | Release date: | 1999-08-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of a penicilloyl-serine transferase of intermediate penicillin sensitivity. The DD-transpeptidase of streptomyces K15.
J.Biol.Chem., 274, 1999
|
|
1UNA
 
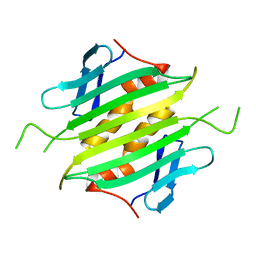 | |
1J9M
 
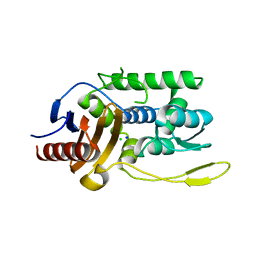 | | K38H mutant of Streptomyces K15 DD-transpeptidase | | Descriptor: | CHLORIDE ION, DD-transpeptidase, SODIUM ION | | Authors: | Fonze, E, Rhazi, N, Nguyen-Disteche, M, Charlier, P. | | Deposit date: | 2001-05-28 | | Release date: | 2001-06-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Catalytic mechanism of the Streptomyces K15 DD-transpeptidase/penicillin-binding protein probed by site-directed mutagenesis and structural analysis.
Biochemistry, 42, 2003
|
|
4DHD
 
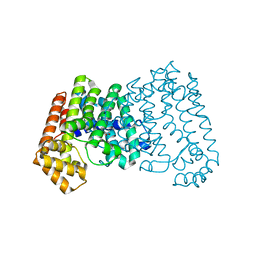 | | Crystal structure of isoprenoid synthase A3MSH1 (TARGET EFI-501992) from Pyrobaculum calidifontis | | Descriptor: | ACETATE ION, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-01-27 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3H0T
 
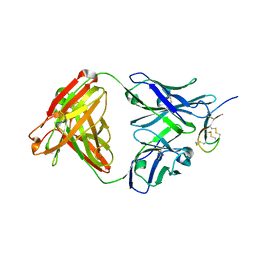 | | Hepcidin-Fab complex | | Descriptor: | Fab fragment, Heavy chain, Light chain, ... | | Authors: | Syed, R, Li, V. | | Deposit date: | 2009-04-10 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Hepcidin revisited, disulfide connectivity, dynamics, and structure.
J.Biol.Chem., 284, 2009
|
|
1ESI
 
 | |
1ES4
 
 | | C98N mutant of streptomyces K15 DD-transpeptidase | | Descriptor: | DD-TRANSPEPTIDASE | | Authors: | Fonze, E, Charlier, P. | | Deposit date: | 2000-04-07 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic mechanism of the Streptomyces K15 DD-transpeptidase/penicillin-binding protein probed by site-directed mutagenesis and structural analysis.
Biochemistry, 42, 2003
|
|
1ES2
 
 | | S96A mutant of streptomyces K15 DD-transpeptidase | | Descriptor: | DD-TRANSPEPTIDASE | | Authors: | Fonze, E, Charlier, P. | | Deposit date: | 2000-04-07 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Catalytic mechanism of the Streptomyces K15 DD-transpeptidase/penicillin-binding protein probed by site-directed mutagenesis and structural analysis.
Biochemistry, 42, 2003
|
|
1ES3
 
 | | C98A mutant of streptomyces K15 DD-transpeptidase | | Descriptor: | DD-TRANSPEPTIDASE, SODIUM ION | | Authors: | Fonze, E, Charlier, P. | | Deposit date: | 2000-04-07 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catalytic mechanism of the Streptomyces K15 DD-transpeptidase/penicillin-binding protein probed by site-directed mutagenesis and structural analysis.
Biochemistry, 42, 2003
|
|
1ES5
 
 | |
3TS7
 
 | | CRYSTAL STRUCTURE OF FARNESYL DIPHOSPHATE SYNTHASE (TARGET EFI-501951) FROM Methylococcus capsulatus | | Descriptor: | Geranyltranstransferase, PHOSPHATE ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-09-12 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3UCA
 
 | | Crystal structure of isoprenoid synthase (target EFI-501974) from clostridium perfringens | | Descriptor: | Geranyltranstransferase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-10-26 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2I7S
 
 | | Crystal structure of Re(phen)(CO)3 (Thr124His)(His83Gln) Azurin Cu(II) from Pseudomonas aeruginosa | | Descriptor: | (1,10 PHENANTHROLINE)-(TRI-CARBON MONOXIDE) RHENIUM (I), Azurin, COBALT TETRAAMMINE ION, ... | | Authors: | Gradinaru, C, Crane, B.R. | | Deposit date: | 2006-08-31 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Relaxation dynamics of Pseudomonas aeruginosa Re(I)(CO)3(alpha-diimine)(HisX)+ (X = 83, 107, 109, 124, 126)Cu(II) azurins.
J.Am.Chem.Soc., 131, 2009
|
|
1XOC
 
 | | The structure of the oligopeptide-binding protein, AppA, from Bacillus subtilis in complex with a nonapeptide. | | Descriptor: | Nonapeptide VDSKNTSSW, Oligopeptide-binding protein appA, ZINC ION | | Authors: | Levdikov, V.M, Blagova, E.V, Brannigan, J.A, Wright, L, Vagin, A.A, Wilkinson, A.J. | | Deposit date: | 2004-10-06 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The structure of the oligopeptide-binding protein, AppA, from Bacillus subtilis in complex with a nonapeptide.
J.Mol.Biol., 345, 2005
|
|
3FW3
 
 | | Crystal Structure of soluble domain of CA4 in complex with Dorzolamide | | Descriptor: | (4S-TRANS)-4-(ETHYLAMINO)-5,6-DIHYDRO-6-METHYL-4H-THIENO(2,3-B)THIOPYRAN-2-SULFONAMIDE-7,7-DIOXIDE, Carbonic anhydrase 4, SULFATE ION, ... | | Authors: | Greasley, S.E, Ferre, R.A.A, Paz, R, Wickersham, J. | | Deposit date: | 2009-01-16 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Thioether benzenesulfonamide inhibitors of carbonic anhydrases II and IV: structure-based drug design, synthesis, and biological evaluation.
Bioorg.Med.Chem., 18, 2010
|
|
2JTX
 
 | |
