3QNF
 
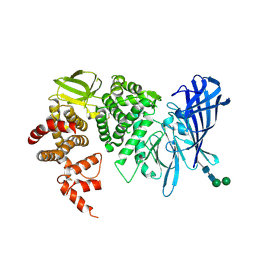 | | Crystal structure of the open state of human endoplasmic reticulum aminopeptidase 1 ERAP1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum aminopeptidase 1, ZINC ION, ... | | Authors: | Vollmar, M, Kochan, G, Krojer, T, Harvey, D, Chaikuad, A, Allerston, C, Muniz, J.R.C, Raynor, J, Ugochukwu, E, Berridge, G, Wordsworth, B.P, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Kavanagh, K, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-02-08 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of the endoplasmic reticulum aminopeptidase-1 (ERAP1) reveal the molecular basis for N-terminal peptide trimming.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4AVP
 
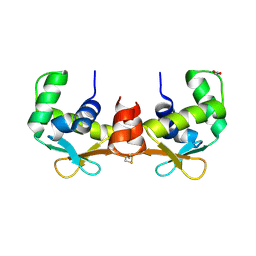 | | Crystal structure of the DNA-binding domain of human ETV1. | | Descriptor: | 1,2-ETHANEDIOL, ETS TRANSLOCATION VARIANT 1 | | Authors: | Allerston, C.K, Cooper, C.D.O, Krojer, T, Chaikuad, A, Filippakopoulos, P, Canning, P, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Gileadi, O. | | Deposit date: | 2012-05-28 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structures of the Ets Domains of Transcription Factors Etv1, Etv4, Etv5 and Fev: Determinants of DNA Binding and Redox Regulation by Disulfide Bond Formation.
J.Biol.Chem., 290, 2015
|
|
3ZHP
 
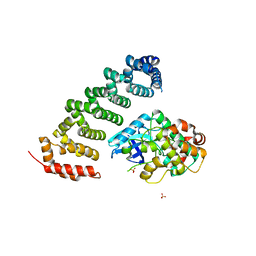 | | Human MST3 (STK24) in complex with MO25beta | | Descriptor: | CALCIUM-BINDING PROTEIN 39-LIKE, SERINE/THREONINE-PROTEIN KINASE 24, SULFATE ION | | Authors: | Elkins, J.M, Szklarz, M, Krojer, T, Mehellou, Y, Alessi, D.R, Chaikaud, A, von Delft, F, Bountra, C, Edwards, A, Knapp, S. | | Deposit date: | 2012-12-24 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Insights Into the Activation of Mst3 by Mo25.
Biochem.Biophys.Res.Commun., 431, 2013
|
|
4AYX
 
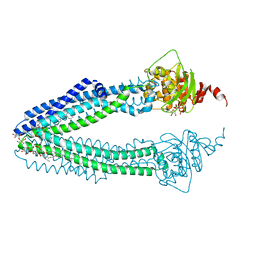 | | STRUCTURE OF THE HUMAN MITOCHONDRIAL ABC TRANSPORTER, ABCB10 (ROD FORM B) | | Descriptor: | ATP-BINDING CASSETTE SUB-FAMILY B MEMBER 10, CARDIOLIPIN, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Pike, A.C.W, Shintre, C.A, Li, Q, Kim, J, von Delft, F, Barr, A.J, Das, S, Chaikuad, A, Xia, X, Quigley, A, Dong, Y, Dong, L, Krojer, T, Vollmar, M, Muniz, J.R.C, Bray, J.E, Berridge, G, Chalk, R, Gileadi, O, Burgess-Brown, N, Shrestha, L, Goubin, S, Yang, J, Mahajan, P, Mukhopadhyay, S, Bullock, A.N, Arrowsmith, C.H, Weigelt, J, Bountra, C, Edwards, A.M, Carpenter, E.P. | | Deposit date: | 2012-06-22 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of Abcb10, a Human ATP-Binding Cassette Transporter in Apo- and Nucleotide-Bound States
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4AYT
 
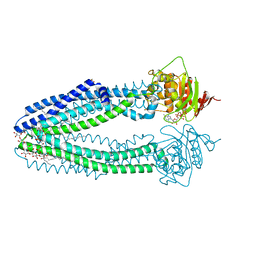 | | STRUCTURE OF THE HUMAN MITOCHONDRIAL ABC TRANSPORTER, ABCB10 | | Descriptor: | ATP-BINDING CASSETTE SUB-FAMILY B MEMBER 10 MITOCHONDRIAL, CARDIOLIPIN, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Pike, A.C.W, Shintre, C.A, Li, Q, Kim, J, von Delft, F, Barr, A.J, Das, S, Chaikuad, A, Xia, X, Quigley, A, Dong, Y, Dong, L, Krojer, T, Vollmar, M, Muniz, J.R.C, Bray, J.E, Berridge, G, Chalk, R, Gileadi, O, Burgess-Brown, N, Shrestha, L, Goubin, S, Yang, J, Mahajan, P, Mukhopadhyay, S, Bullock, A.N, Arrowsmith, C.H, Weigelt, J, Bountra, C, Edwards, A.M, Carpenter, E.P. | | Deposit date: | 2012-06-22 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structures of Abcb10, a Human ATP-Binding Cassette Transporter in Apo- and Nucleotide-Bound States
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4AGU
 
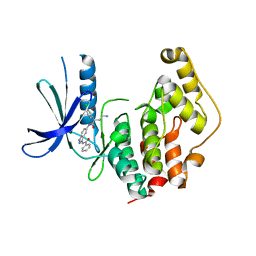 | | CRYSTAL STRUCTURE OF THE HUMAN CDKL1 KINASE DOMAIN | | Descriptor: | CYCLIN-DEPENDENT KINASE-LIKE 1, N-(5-{[(2S)-4-amino-2-(3-chlorophenyl)butanoyl]amino}-1H-indazol-3-yl)benzamide | | Authors: | Canning, P, Sharpe, T.D, Allerston, C, Savitsky, P, Pike, A.C.W, Muniz, J.R.C, Chaikuad, A, Kuo, K, Burgess-Brown, N, Ayinampudi, V, Zhang, Y, Thangaratnarajah, C, Ugochukwu, E, Vollmar, M, Krojer, T, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Knapp, S, Bullock, A. | | Deposit date: | 2012-01-31 | | Release date: | 2012-02-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CDKL Family Kinases Have Evolved Distinct Structural Features and Ciliary Function.
Cell Rep, 22, 2018
|
|
4BNC
 
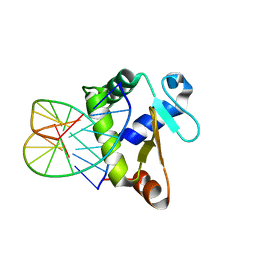 | | Crystal structure of the DNA-binding domain of human ETV1 complexed with DNA | | Descriptor: | 5'-D(*AP*CP*CP*GP*GP*AP*AP*GP*TP*GP)-3', 5'-D(*CP*AP*CP*TP*TP*CP*CP*GP*GP*TP)-3', HUMAN ETV1 | | Authors: | Allerston, C.K, Cooper, C.D.O, Krojer, T, Chaikuad, A, Vollmar, M, Froese, D.S, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Gileadi, O. | | Deposit date: | 2013-05-14 | | Release date: | 2013-07-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the Ets Domains of Transcription Factors Etv1, Etv4, Etv5 and Fev: Determinants of DNA Binding and Redox Regulation by Disulfide Bond Formation.
J.Biol.Chem., 290, 2015
|
|
4APF
 
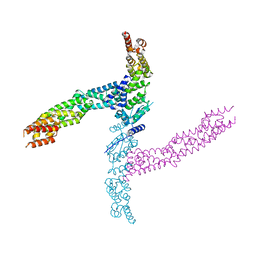 | | Crystal structure of the human KLHL11-Cul3 complex at 3.1A resolution | | Descriptor: | CULLIN 3, KELCH-LIKE PROTEIN 11 | | Authors: | Canning, P, Cooper, C.D.O, Krojer, T, Vollmar, M, Ugochukwu, E, Muniz, J.R.C, Ayinampudi, V, Savitsky, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Bullock, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-04-02 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for Cul3 Assembly with the Btb-Kelch Family of E3 Ubiquitin Ligases.
J.Biol.Chem., 288, 2013
|
|
4AP2
 
 | | Crystal structure of the human KLHL11-Cul3 complex at 2.8A resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CULLIN-3, ... | | Authors: | Canning, P, Cooper, C.D.O, Krojer, T, Filippakopoulos, P, Ayinampudi, V, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Bullock, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-03-30 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Cul3 Assembly with the Btb-Kelch Family of E3 Ubiquitin Ligases.
J.Biol.Chem., 288, 2013
|
|
4ASC
 
 | | Crystal structure of the Kelch domain of human KBTBD5 | | Descriptor: | 1,2-ETHANEDIOL, KELCH REPEAT AND BTB DOMAIN-CONTAINING PROTEIN 5 | | Authors: | Canning, P, Ayinampudi, V, Krojer, T, Strain-Damerell, C, Raynor, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-04-30 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Basis for Cul3 Assembly with the Btb-Kelch Family of E3 Ubiquitin Ligases.
J.Biol.Chem., 288, 2013
|
|
2XN4
 
 | | Crystal structure of the kelch domain of human KLHL2 (Mayven) | | Descriptor: | 1,2-ETHANEDIOL, KELCH-LIKE PROTEIN 2, SODIUM ION, ... | | Authors: | Canning, P, Hozjan, V, Cooper, C.D.O, Ayinampudi, V, Vollmar, M, Pike, A.C.W, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2010-07-30 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Basis for Cul3 Assembly with the Btb-Kelch Family of E3 Ubiquitin Ligases.
J.Biol.Chem., 288, 2013
|
|
2YDY
 
 | | Crystal structure of human S-adenosylmethionine synthetase 2, beta subunit in Orthorhombic crystal form | | Descriptor: | CHLORIDE ION, METHIONINE ADENOSYLTRANSFERASE 2 SUBUNIT BETA, SULFATE ION | | Authors: | Yue, W.W, Shafqat, N, Muniz, J.R.C, Pike, A.C.W, Chaikuad, A, Allerston, C.K, Gileadi, O, von Delft, F, Kavanagh, K.L, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Oppermann, U. | | Deposit date: | 2011-03-25 | | Release date: | 2011-04-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Insight Into S-Adenosylmethionine Biosynthesis from the Crystal Structures of the Human Methionine Adenosyltransferase Catalytic and Regulatory Subunits.
Biochem.J., 452, 2013
|
|
2WZK
 
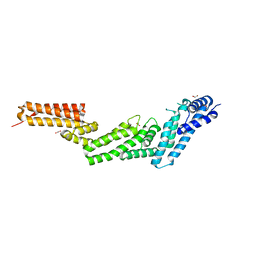 | | Structure of the Cul5 N-terminal domain at 2.05A resolution. | | Descriptor: | 1,2-ETHANEDIOL, CULLIN-5 | | Authors: | Muniz, J.R.C, Ayinampudi, V, Zhang, Y, Babon, J.J, Chaikuad, A, Krojer, T, Pike, A.C.W, Vollmar, M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Bullock, A.N. | | Deposit date: | 2009-11-30 | | Release date: | 2009-12-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular Architecture of the Ankyrin Socs Box Family of Cul5-Dependent E3 Ubiquitin Ligases
J.Mol.Biol., 425, 2013
|
|
3K2O
 
 | | Structure of an oxygenase | | Descriptor: | ACETATE ION, Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, CHLORIDE ION, ... | | Authors: | Krojer, T, McDonough, M.A, Clifton, I.J, Mantri, M, Ng, S.S, Pike, A.C.W, Butler, D.S, Webby, C.J, Kochan, G, Bhatia, C, Bray, J.E, Chaikuad, A, Gileadi, O, von Delft, F, Weigelt, J, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Schofield, C.J, Kavanagh, K.L, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-30 | | Release date: | 2009-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the 2-Oxoglutarate- and Fe(II)-Dependent Lysyl Hydroxylase JMJD6.
J.Mol.Biol., 401, 2010
|
|
3MQM
 
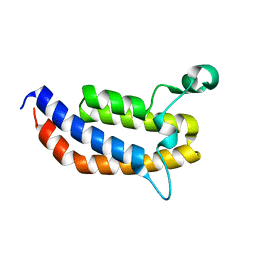 | | Crystal Structure of the Bromodomain of human ASH1L | | Descriptor: | Probable histone-lysine N-methyltransferase ASH1L | | Authors: | Filippakopoulos, P, Picaud, S, Keates, T, Felletar, I, Vollmar, M, Chaikuad, A, Krojer, T, Canning, P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-28 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
1ANJ
 
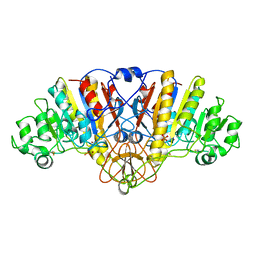 | | ALKALINE PHOSPHATASE (K328H) | | Descriptor: | ALKALINE PHOSPHATASE, PHOSPHATE ION, ZINC ION | | Authors: | Murphy, J.E, Tibbitts, T.T, Kantrowitz, E.R. | | Deposit date: | 1995-09-06 | | Release date: | 1996-01-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutations at positions 153 and 328 in Escherichia coli alkaline phosphatase provide insight towards the structure and function of mammalian and yeast alkaline phosphatases.
J.Mol.Biol., 253, 1995
|
|
1ANI
 
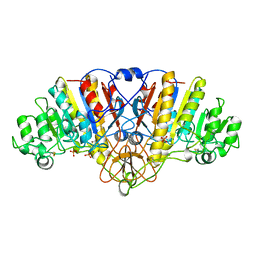 | | ALKALINE PHOSPHATASE (D153H, K328H) | | Descriptor: | ALKALINE PHOSPHATASE, PHOSPHATE ION, ZINC ION | | Authors: | Murphy, J.E, Tibbitts, T.T, Kantrowitz, E.R. | | Deposit date: | 1995-09-06 | | Release date: | 1996-01-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutations at positions 153 and 328 in Escherichia coli alkaline phosphatase provide insight towards the structure and function of mammalian and yeast alkaline phosphatases.
J.Mol.Biol., 253, 1995
|
|
