2VX1
 
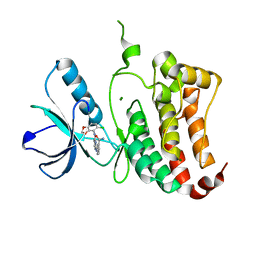 | | ephB4 kinase domain inhibitor complex | | Descriptor: | 3-({4-[(5-CHLORO-1,3-BENZODIOXOL-4-YL)AMINO]PYRIMIDIN-2-YL}AMINO)BENZAMIDE, EPHRIN TYPE-B RECEPTOR 4, MAGNESIUM ION | | Authors: | Read, J, Brassington, C.A, Green, I, McCall, E.J, Valentine, A.L, Barratt, D, Leach, A.G, Kettle, J.G. | | Deposit date: | 2008-06-30 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Inhibitors of the Tyrosine Kinase Ephb4. Part 2: Structure-Based Discovery and Optimisation of 3,5-Bis Substituted Anilinopyrimidines.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3DOW
 
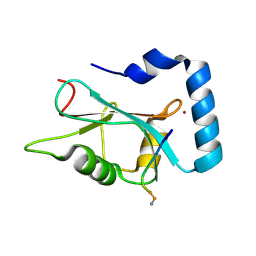 | |
6CN3
 
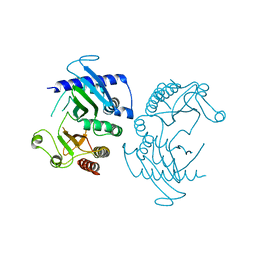 | |
7BN3
 
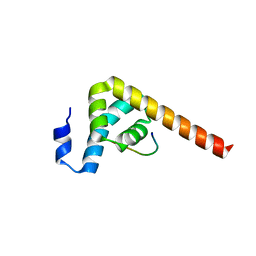 | |
7BN1
 
 | |
7BH8
 
 | | 3H4-Fab HLA-E-VL9 co-complex | | Descriptor: | 3H4 Fab heavy chain, 3H4 Fab light chain, Beta-2-microglobulin, ... | | Authors: | Walters, L.C, Rozbesky, D. | | Deposit date: | 2021-01-10 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mouse and human antibodies bind HLA-E-leader peptide complexes and enhance NK cell cytotoxicity.
Commun Biol, 5, 2022
|
|
3ZUL
 
 | |
1M5Q
 
 | | Crystal structure of a novel Sm-like archaeal protein from Pyrobaculum aerophilum | | Descriptor: | ACETIC ACID, CADMIUM ION, GLYCEROL, ... | | Authors: | Mura, C, Phillips, M, Kozhukhovsky, A, Eisenberg, D. | | Deposit date: | 2002-07-09 | | Release date: | 2003-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and assembly of an augmented Sm-like archaeal protein 14-mer
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
2A6E
 
 | | Crystal structure of the T. Thermophilus RNA polymerase holoenzyme | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Artsimovitch, I, Vassylyeva, M.N, Svetlov, D, Svetlov, V, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Tahirov, T.H, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-02 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins.
Cell(Cambridge,Mass.), 122, 2005
|
|
8P4W
 
 | | Lactobacillus plantarum LpdD mutant - H35A | | Descriptor: | PHOSPHATE ION, Protein LpdD | | Authors: | Gahloth, D, Leys, D. | | Deposit date: | 2023-05-23 | | Release date: | 2024-06-12 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The prFMNH 2 -binding chaperone LpdD assists UbiD decarboxylase activation.
J.Biol.Chem., 300, 2024
|
|
3HG0
 
 | | Crystal structure of a DARPin in complex with ORF49 from Lactococcal phage TP901-1 | | Descriptor: | Baseplate protein, Designed Ankyrin Repeat Protein (DARPin) 20 | | Authors: | Veesler, D, Dreier, B, Blangy, S, Lichiere, J, Tremblay, D, Moineau, S, Spinelli, S, Tegoni, M, Pluckthun, A, Campanacci, V, Cambillau, C. | | Deposit date: | 2009-05-13 | | Release date: | 2009-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and function of a DARPin neutralizing inhibitor of lactococcal phage TP901-1: comparison of DARPin and camelid VHH binding mode.
J.Biol.Chem., 284, 2009
|
|
8S53
 
 | | X-ray crystal structure of CYP142 from Mycobacterium tuberculosis in complex with a fragment bound in two poses | | Descriptor: | 4-benzylpyridine, PROTOPORPHYRIN IX CONTAINING FE, Steroid C26-monooxygenase | | Authors: | Snee, M, Kavanagh, M, Levy, C, Leys, D. | | Deposit date: | 2024-02-22 | | Release date: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fragment-based development of small molecule inhibitors targeting Mycobacterium tuberculosis cholesterol metabolism.
Biorxiv, 2024
|
|
6CGO
 
 | | Molecular basis for condensation domain-mediated chain release from the enacyloxin polyketide synthase | | Descriptor: | Condensation domain protein, PHOSPHATE ION | | Authors: | Valentic, T.R, Tsai, S.C, Challis, G.L, Lewandowski, J.R, Kosol, S, Gallo, A, Griffiths, D, Masschelein, J.L, Jenner, M, De los Santos, E. | | Deposit date: | 2018-02-20 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for chain release from the enacyloxin polyketide synthase.
Nat.Chem., 11, 2019
|
|
6R8B
 
 | | Escherichia coli AGPase in complex with FBP. | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Glucose-1-phosphate adenylyltransferase | | Authors: | Cifuente, J.O, Comino, N, D'Angelo, C, Marina, A, Gil-Carton, D, Albesa-Jove, D, Guerin, M.E. | | Deposit date: | 2019-04-01 | | Release date: | 2020-02-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The allosteric control mechanism of bacterial glycogen biosynthesis disclosed by cryoEM
Biorxiv, 2020
|
|
6R8U
 
 | | Escherichia coli AGPase in complex with AMP. | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glucose-1-phosphate adenylyltransferase | | Authors: | Cifuente, J.O, Comino, N, D'Angelo, C, Marina, A, Gil-Carton, D, Albesa-Jove, D, Guerin, M.E. | | Deposit date: | 2019-04-02 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The allosteric control mechanism of bacterial glycogen biosynthesis disclosed by cryoEM
Biorxiv, 2020
|
|
1UN6
 
 | | THE CRYSTAL STRUCTURE OF A ZINC FINGER - RNA COMPLEX REVEALS TWO MODES OF MOLECULAR RECOGNITION | | Descriptor: | 5S RIBOSOMAL RNA, MAGNESIUM ION, TRANSCRIPTION FACTOR IIIA, ... | | Authors: | Lu, D, Searles, M.A, Klug, A. | | Deposit date: | 2003-09-04 | | Release date: | 2003-11-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of a Zinc-Finger-RNA Complex Reveals Two Modes of Molecular Recognition
Nature, 426, 2003
|
|
4QE8
 
 | | FXR with DM175 and NCoA-2 peptide | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,2-ETHANEDIOL, 4-({2-[(4-tert-butylbenzoyl)amino]benzoyl}amino)benzoic acid, ... | | Authors: | Kudlinzki, D, Merk, D, Linhard, V.L, Saxena, K, Sreeramulu, S, Nilsson, E, Dekker, N, Wissler, L, Bamberg, K, Schubert-Zsilavecz, M, Schwalbe, H. | | Deposit date: | 2014-05-15 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | FXR with DM175 and NCoA-2 peptide
To be Published
|
|
4MHA
 
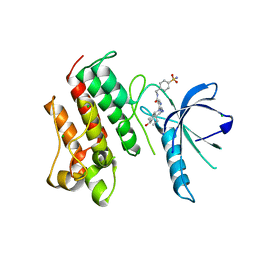 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC1817 | | Descriptor: | 2-(butylamino)-4-[(trans-4-hydroxycyclohexyl)amino]-N-(4-sulfamoylbenzyl)pyrimidine-5-carboxamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Zhang, W, Mciver, A, Stashko, M.A, Deryckere, D, Branchford, B.R, Hunter, D, Kireev, D.B, Miley, D.B.M, Norris-Drouin, J, Stewart, W.M, Lee, M, Sather, S, Zhou, Y, Dipaola, J.A, Machius, M, Janzen, W.P, Earp, H.S, Graham, D.K, Frye, S, Wang, X. | | Deposit date: | 2013-08-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Discovery of Mer specific tyrosine kinase inhibitors for the treatment and prevention of thrombosis.
J.Med.Chem., 56, 2013
|
|
7CR2
 
 | | human KCNQ2 in complex with retigabine | | Descriptor: | Potassium voltage-gated channel subfamily KQT member 2, ethyl N-[2-azanyl-4-[(4-fluorophenyl)methylamino]phenyl]carbamate | | Authors: | Li, X, Lv, D, Wang, J, Ye, S, Guo, J. | | Deposit date: | 2020-08-12 | | Release date: | 2020-09-16 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis for ligand activation of the human KCNQ2 channel.
Cell Res., 31, 2021
|
|
7YWX
 
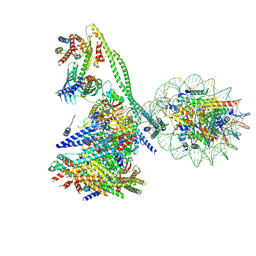 | | Structure of the human CCAN CENP-A alpha-satellite complex | | Descriptor: | Centromere protein C, Centromere protein H, Centromere protein I, ... | | Authors: | Yatskevich, S, Muir, K.W, Bellini, D, Zhang, Z, Yang, J, Tischer, T, Predin, M, Dendooven, T, McLaughlin, S.H, Barford, D. | | Deposit date: | 2022-02-14 | | Release date: | 2022-05-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome.
Science, 376, 2022
|
|
6P0T
 
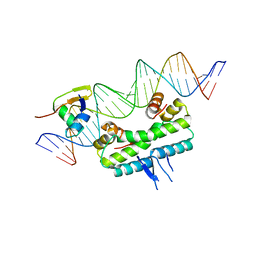 | | Crystal structure of ternary DNA complex "FX(1-2)-1Xis" containing E. coli Fis and phage lambda Xis | | Descriptor: | DNA (27-MER), FX1-2, DNA-binding protein Fis, ... | | Authors: | Hancock, S.P, Cascio, D, Johnson, R.C. | | Deposit date: | 2019-05-17 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.603 Å) | | Cite: | Cooperative DNA binding by proteins through DNA shape complementarity.
Nucleic Acids Res., 47, 2019
|
|
7YYH
 
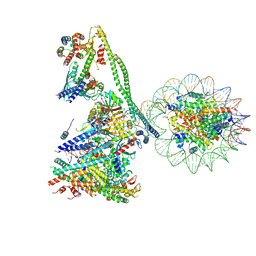 | | Structure of the human CCANdeltaT CENP-A alpha-satellite complex | | Descriptor: | Centromere protein C, Centromere protein H, Centromere protein I, ... | | Authors: | Yatskevich, S, Muir, K.W, Bellini, D, Zhang, Z, Yang, J, Tischer, T, Predin, M, Dendooven, T, McLaughlin, S.H, Barford, D. | | Deposit date: | 2022-02-17 | | Release date: | 2022-05-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome.
Science, 376, 2022
|
|
8OVD
 
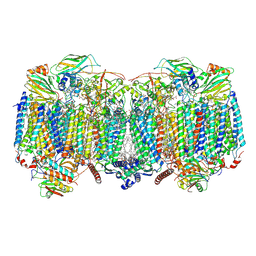 | | Respiratory supercomplex (III2-IV2) from Mycobacterium smegmatis | | Descriptor: | (1R)-2-(dodecanoyloxy)-1-[(phosphonooxy)methyl]ethyl tetradecanoate, (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2S)-1-(hexadecanoyloxy)propan-2-yl (10S)-10-methyloctadecanoate, ... | | Authors: | Kovalova, T, Krol, S, Sjostrand, D, Riepl, D, Gamiz-Hernandez, A, Brzezinski, P, Kaila, V, Hogbom, M. | | Deposit date: | 2023-04-25 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Long-range charge transfer mechanism of the III 2 IV 2 mycobacterial supercomplex.
Nat Commun, 15, 2024
|
|
8OVC
 
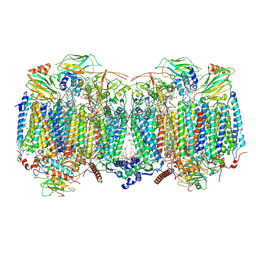 | | Respiratory supercomplex (III2-IV2) from Mycobacterium smegmatis | | Descriptor: | (1R)-2-(dodecanoyloxy)-1-[(phosphonooxy)methyl]ethyl tetradecanoate, (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2S)-1-(hexadecanoyloxy)propan-2-yl (10S)-10-methyloctadecanoate, ... | | Authors: | Kovalova, T, Krol, S, Sjostrand, D, Riepl, D, Gamiz-Hernandez, A, Brzezinski, P, Kaila, V, Hogbom, M. | | Deposit date: | 2023-04-25 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Long-range charge transfer mechanism of the III 2 IV 2 mycobacterial supercomplex.
Nat Commun, 15, 2024
|
|
6RBD
 
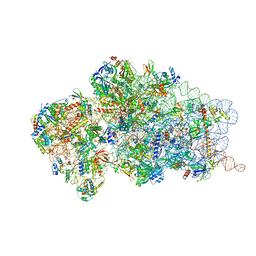 | | State 1 of yeast Tsr1-TAP Rps20-Deltaloop pre-40S particles | | Descriptor: | 20S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Shayan, R, Mitterer, V, Ferreira-Cerca, S, Murat, G, Enne, T, Rinaldi, D, Weigl, S, Omanic, H, Gleizes, P.E, Kressler, D, Pertschy, B, Plisson-Chastang, C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Conformational proofreading of distant 40S ribosomal subunit maturation events by a long-range communication mechanism.
Nat Commun, 10, 2019
|
|
