7E0B
 
 | |
7E3J
 
 | |
7D72
 
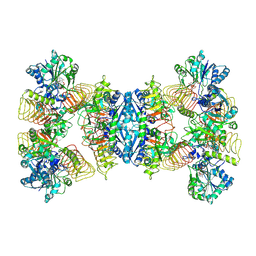 | | Cryo-EM structures of human GMPPA/GMPPB complex bound to GDP-Mannose | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, MAGNESIUM ION, Mannose-1-phosphate guanyltransferase alpha, ... | | Authors: | Zheng, L, Liu, Z, Wang, Y, Yang, F, Wang, J, Qing, J, Cai, X, Mo, X, Gao, N, Jia, D. | | Deposit date: | 2020-10-02 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of human GMPPA-GMPPB complex reveal how cells maintain GDP-mannose homeostasis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7D73
 
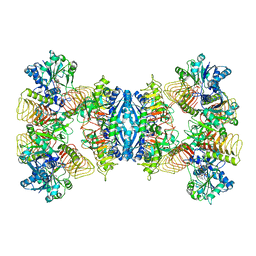 | | Cryo-EM structure of GMPPA/GMPPB complex bound to GTP (State I) | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, GUANOSINE-5'-TRIPHOSPHATE, Mannose-1-phosphate guanyltransferase alpha, ... | | Authors: | Zheng, L, Liu, Z, Wang, Y, Yang, F, Wang, J, Qing, J, Cai, X, Mo, X, Gao, N, Jia, D. | | Deposit date: | 2020-10-02 | | Release date: | 2021-05-05 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of human GMPPA-GMPPB complex reveal how cells maintain GDP-mannose homeostasis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7D74
 
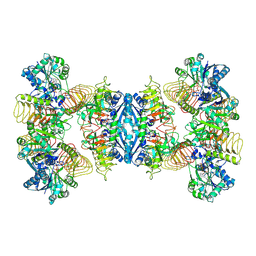 | | Cryo-EM structure of GMPPA/GMPPB complex bound to GTP (state II) | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Mannose-1-phosphate guanyltransferase alpha, Mannose-1-phosphate guanyltransferase beta | | Authors: | Zheng, L, Liu, Z, Wang, Y, Yang, F, Wang, J, Qing, J, cai, X, Mo, X, Gao, N, Jia, D. | | Deposit date: | 2020-10-02 | | Release date: | 2021-05-19 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of human GMPPA-GMPPB complex reveal how cells maintain GDP-mannose homeostasis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
5EQR
 
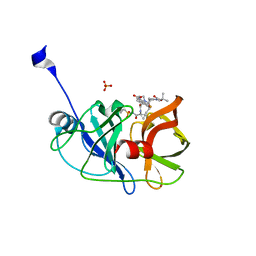 | | Crystal structure of a genotype 1a/3a chimeric HCV NS3/4A protease in complex with danoprevir | | Descriptor: | (2R,6S,12Z,13aS,14aR,16aS)-6-[(tert-butoxycarbonyl)amino]-14a-[(cyclopropylsulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8 ,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-2-yl 4-fluoro-2H-isoindole-2-carboxylate, NS3 protease, SULFATE ION, ... | | Authors: | Soumana, D, Yilmaz, N.K, Ali, A, Prachanronarong, K.L, Schiffer, C.A. | | Deposit date: | 2015-11-13 | | Release date: | 2016-11-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Molecular and Dynamic Mechanism Underlying Drug Resistance in Genotype 3 Hepatitis C NS3/4A Protease.
J.Am.Chem.Soc., 138, 2016
|
|
7EY5
 
 | |
7EYA
 
 | |
7EY0
 
 | |
7EZV
 
 | |
7EY4
 
 | | Local CryoEM of the SARS-CoV-2 S6PV2 in complex with BD-667 | | Descriptor: | BD-667 H, BD-667 L, Spike glycoprotein, ... | | Authors: | Liu, P.L. | | Deposit date: | 2021-05-29 | | Release date: | 2021-09-08 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Structures of SARS-CoV-2 B.1.351 neutralizing antibodies provide insights into cocktail design against concerning variants.
Cell Res., 31, 2021
|
|
5ESB
 
 | | Crystal structure of a genotype 1a/3a chimeric HCV NS3/4A protease in complex with Vaniprevir | | Descriptor: | (5R,7S,10S)-10-tert-butyl-N-{(1R,2R)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethylcyclopropyl}-15,15-dimethyl-3,9,12-trioxo-6,7,9,10,11,12,14,15,16,17,18,19-dodecahydro-1H,5H-2,23:5,8-dimethano-4,13,2,8,11-benzodioxatriazacyclohenicosine-7(3H)-carboxamide, NS3 protease, SULFATE ION, ... | | Authors: | Soumana, D, Yilmaz, N.K, Ali, A, Prachanronarong, K.L, Schiffer, C.A. | | Deposit date: | 2015-11-16 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular and Dynamic Mechanism Underlying Drug Resistance in Genotype 3 Hepatitis C NS3/4A Protease.
J.Am.Chem.Soc., 138, 2016
|
|
5EPY
 
 | | Crystal structure of HCV NS3/4A protease A156T variant in complex with 5172-mcP1P3 (MK-5172 P1-P3 macrocyclic analogue) | | Descriptor: | 2-Methyl-2-propanyl {(2R,6S,12Z,13aS,14aR,16aS)-14a-[(cyclopropylsulfonyl)carbamoyl]-2-[(3-ethyl-7-methoxy-2-quinoxalinyl)oxy]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclop ropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, NS3 protease, SULFATE ION, ... | | Authors: | Soumana, D.I, Yilmaz, N.K, Ali, A, Prachanronarong, K.L, Aydin, C, Schiffer, C.A. | | Deposit date: | 2015-11-12 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Thermodynamic Effects of Macrocyclization in HCV NS3/4A Inhibitor MK-5172.
Acs Chem.Biol., 11, 2016
|
|
5EQQ
 
 | | Crystal structure of HCV NS3/4A WT protease in complex with 5172-Linear (MK-5172 linear analogue) | | Descriptor: | NS3 protease, SULFATE ION, ZINC ION, ... | | Authors: | Soumana, D, Yilmaz, N.K, Ali, A, Prachanronarong, K.L, Aydin, C, Schiffer, C.A. | | Deposit date: | 2015-11-13 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Thermodynamic Effects of Macrocyclization in HCV NS3/4A Inhibitor MK-5172.
Acs Chem.Biol., 11, 2016
|
|
5EPN
 
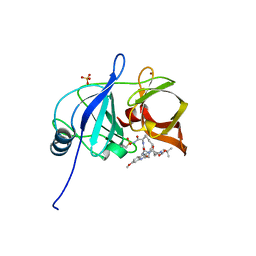 | | Crystal structure of HCV NS3/4A protease in complex with 5172-mcP1P3 (MK-5172 P1-P3 macrocyclic analogue) | | Descriptor: | 2-Methyl-2-propanyl {(2R,6S,12Z,13aS,14aR,16aS)-14a-[(cyclopropylsulfonyl)carbamoyl]-2-[(3-ethyl-7-methoxy-2-quinoxalinyl)oxy]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclop ropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, NS3 protease, SULFATE ION, ... | | Authors: | Soumana, D.I, Yilmaz, N.K, Ali, A, Prachanronarong, K.L, Aydin, C, Schiffer, C.A. | | Deposit date: | 2015-11-11 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Thermodynamic Effects of Macrocyclization in HCV NS3/4A Inhibitor MK-5172.
Acs Chem.Biol., 11, 2016
|
|
7CAC
 
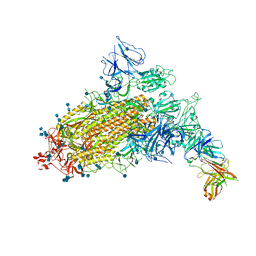 | | SARS-CoV-2 S trimer with one RBD in the open state and complexed with one H014 Fab. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H014 Fab, ... | | Authors: | Zhe, L, Cao, L, Deng, Y, Sun, Y, Wang, N, Xie, L, Wang, Y, Rao, Z, Qin, C, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2021-02-24 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody.
Science, 369, 2020
|
|
7CAH
 
 | | The interface of H014 Fab binds to SARS-CoV-2 S | | Descriptor: | Heavy chain of H014 Fab, Light chain of H014 Fab, Spike protein S1 | | Authors: | Zhe, L, Cao, L, Deng, Y, Sun, Y, Wang, N, Xie, L, Rao, Z, wang, Y, Qin, C, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2020-08-12 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody.
Science, 369, 2020
|
|
7CHS
 
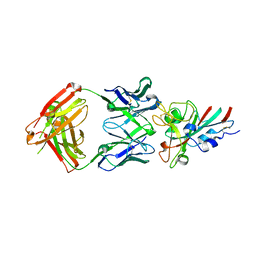 | | Crystal structure of SARS-CoV-2 antibody P22A-1D1 with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P22A-1D1 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Ge, J, Wang, R, Zhang, Q. | | Deposit date: | 2020-07-06 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Potent and protective IGHV3-53/3-66 public antibodies and their shared escape mutant on the spike of SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7CHO
 
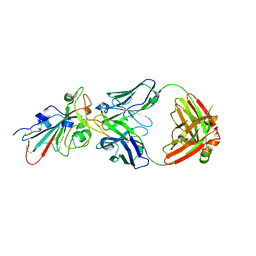 | | Crystal structure of SARS-CoV-2 antibody P5A-1D2 with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P5A-1D2 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Ge, J, Wang, R, Zhang, Q. | | Deposit date: | 2020-07-06 | | Release date: | 2021-05-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.561 Å) | | Cite: | Potent and protective IGHV3-53/3-66 public antibodies and their shared escape mutant on the spike of SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7CHP
 
 | | Crystal structure of SARS-CoV-2 antibody P5A-3C8 with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P5A-3C8 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Ge, J, Wang, R, Zhang, Q. | | Deposit date: | 2020-07-06 | | Release date: | 2021-05-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.357 Å) | | Cite: | Potent and protective IGHV3-53/3-66 public antibodies and their shared escape mutant on the spike of SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7CAK
 
 | | SARS-CoV-2 S trimer with three RBD in the open state and complexed with three H014 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H014 Fab, ... | | Authors: | Zhe, L, Cao, L, Deng, Y, Sun, Y, Wang, N, Xie, L, Wang, Y, Rao, Z, Qin, C, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2020-09-23 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody.
Science, 369, 2020
|
|
7CAB
 
 | | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhe, L, Cao, L, Deng, Y, Sun, Y, Wang, N, Xie, L, Wang, Y, Rao, Z, Qin, C, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody.
Science, 369, 2020
|
|
7L7P
 
 | |
7L7O
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR04-49 | | Descriptor: | (1R,3r,5S)-bicyclo[3.1.0]hexan-3-yl [(2R,6S,12Z,13aS,14aR,16aS)-2-{[6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl]oxy}-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, 1,2-ETHANEDIOL, NS3/4A protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2020-12-29 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of Quinoxaline-Based P1-P3 Macrocyclic NS3/4A Protease Inhibitors with Potent Activity against Drug-Resistant Hepatitis C Virus Variants.
J.Med.Chem., 64, 2021
|
|
7L7N
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR02-59 | | Descriptor: | 1,2-ETHANEDIOL, 1-(trifluoromethyl)cyclobutyl [(2R,6S,12Z,13aS,14aR,16aS)-2-{[6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl]oxy}-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, NS3 protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2020-12-29 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Discovery of Quinoxaline-Based P1-P3 Macrocyclic NS3/4A Protease Inhibitors with Potent Activity against Drug-Resistant Hepatitis C Virus Variants.
J.Med.Chem., 64, 2021
|
|
