1MI0
 
 | | Crystal Structure of the redesigned protein G variant NuG2 | | Descriptor: | immunoglobulin-binding protein G | | Authors: | Nauli, S, Kuhlman, B, Le Trong, I, Stenkamp, R.E, Teller, D.C, Baker, D. | | Deposit date: | 2002-08-21 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures and increased stabilization of the protein G variants with switched folding pathways NuG1 and NuG2
Biochemistry, 11, 2002
|
|
4KYZ
 
 | | Three-dimensional structure of triclinic form of de novo design insertion domain, Northeast Structural Genomics Consortium (NESG) Target OR327 | | Descriptor: | Designed protein OR327 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Maglaqui, M, Xiao, R, Lee, D, Gleixner, J, Baker, D, Everett, J.K, Acton, T.B, Kornhaber, G, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-29 | | Release date: | 2013-07-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.492 Å) | | Cite: | Precise assembly of complex beta sheet topologies from de novo designed building blocks.
Elife, 4, 2015
|
|
2LCB
 
 | | Solution Structure of a Minor and Transiently Formed State of a T4 Lysozyme Mutant | | Descriptor: | Lysozyme | | Authors: | Bouvignies, G, Vallurupalli, P, Hansen, D, Correia, B, Lange, O, Bah, A, Vernon, R.M, Dahlquist, F.W, Baker, D, Kay, L.E. | | Deposit date: | 2011-04-26 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a minor and transiently formed state of a T4 lysozyme mutant.
Nature, 477, 2011
|
|
2LC9
 
 | | Solution Structure of a Minor and Transiently Formed State of a T4 Lysozyme Mutant | | Descriptor: | Lysozyme | | Authors: | Bouvignies, G, Vallurupalli, P, Hansen, D, Correia, B, Lange, O, Bah, A, Vernon, R.M, Dahlquist, F.W, Baker, D, Kay, L.E. | | Deposit date: | 2011-04-26 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a minor and transiently formed state of a T4 lysozyme mutant.
Nature, 477, 2011
|
|
4KY3
 
 | | Three-dimensional Structure of the orthorhombic crystal of computationally designed insertion domain , Northeast Structural Genomics Consortium (NESG) Target OR327 | | Descriptor: | designed protein OR327 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Maglaqui, M, Xiao, R, Lee, D, Gleixner, J, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-28 | | Release date: | 2013-06-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.964 Å) | | Cite: | Precise assembly of complex beta sheet topologies from de novo designed building blocks.
Elife, 4, 2015
|
|
2ERH
 
 | | Crystal Structure of the E7_G/Im7_G complex; a designed interface between the colicin E7 DNAse and the Im7 immunity protein | | Descriptor: | Colicin E7, Colicin E7 immunity protein | | Authors: | Joachimiak, L.A, Kortemme, T, Stoddard, B.L, Baker, D. | | Deposit date: | 2005-10-24 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational Design of a New Hydrogen Bond Network and at Least a 300-fold Specificity Switch at a Protein-Protein Interface.
J.Mol.Biol., 361, 2006
|
|
2GJ7
 
 | | Crystal Structure of a gE-gI/Fc complex | | Descriptor: | Glycoprotein E, Ig gamma-1 chain C region, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Sprague, E.R, Wang, C, Baker, D, Bjorkman, P.J. | | Deposit date: | 2006-03-30 | | Release date: | 2006-05-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Crystal Structure of the HSV-1 Fc Receptor Bound to Fc Reveals a Mechanism for Antibody Bipolar Bridging.
Plos Biol., 4, 2006
|
|
3J02
 
 | | Lidless D386A Mm-cpn in the pre-hydrolysis ATP-bound state | | Descriptor: | Lidless D386A Mm-cpn variant | | Authors: | Zhang, J, Ma, B, DiMaio, F, Douglas, N.R, Joachimiak, L, Baker, D, Frydman, J, Levitt, M, Chiu, W. | | Deposit date: | 2011-02-10 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Cryo-EM structure of a group II chaperonin in the prehydrolysis ATP-bound state leading to lid closure.
Structure, 19, 2011
|
|
5TZO
 
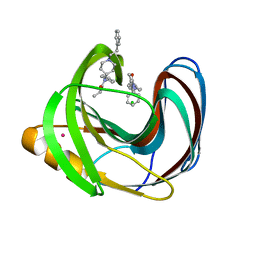 | | Computationally Designed Fentanyl Binder - Fen49*-Complex | | Descriptor: | CHLORIDE ION, Endo-1,4-beta-xylanase A, N-phenyl-N-[1-(2-phenylethyl)piperidin-4-yl]propanamide, ... | | Authors: | Bick, M.J, Greisen, P.J, Morey, K.J, Antunes, M.S, La, D, Sankaran, B, Reymond, L, Johnsson, K, Medford, J.I, Baker, D. | | Deposit date: | 2016-11-22 | | Release date: | 2017-10-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Computational design of environmental sensors for the potent opioid fentanyl.
Elife, 6, 2017
|
|
3J1X
 
 | | A refined model of the prototypical Salmonella typhimurium T3SS basal body reveals the molecular basis for its assembly | | Descriptor: | Protein PrgH | | Authors: | Sgourakis, N.G, Bergeron, J.R.C, Worrall, L.J, Strynadka, N.C.J, Baker, D. | | Deposit date: | 2012-07-10 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.7 Å) | | Cite: | A Refined Model of the Prototypical Salmonella SPI-1 T3SS Basal Body Reveals the Molecular Basis for Its Assembly.
Plos Pathog., 9, 2013
|
|
3J6E
 
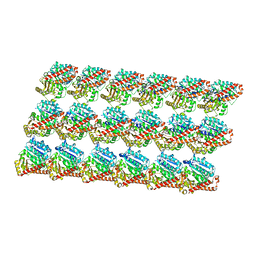 | | Energy minimized average structure of Microtubules stabilized by GmpCpp | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Alushin, G.M, Lander, G.C, Kellogg, E.H, Zhang, R, Baker, D, Nogales, E. | | Deposit date: | 2014-02-18 | | Release date: | 2014-06-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | High-Resolution Microtubule Structures Reveal the Structural Transitions in alpha beta-Tubulin upon GTP Hydrolysis.
Cell(Cambridge,Mass.), 157, 2014
|
|
3J89
 
 | | Structural Plasticity of Helical Nanotubes Based on Coiled-Coil Assemblies | | Descriptor: | synthetic peptide | | Authors: | Egelman, E.H, Xu, C, DiMaio, F, Magnotti, E, Modlin, C, Yu, X, Wright, E, Baker, D, Conticello, V.P. | | Deposit date: | 2014-10-07 | | Release date: | 2015-02-11 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural plasticity of helical nanotubes based on coiled-coil assemblies.
Structure, 23, 2015
|
|
3J6F
 
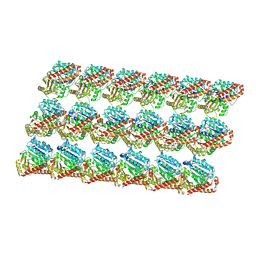 | | Minimized average structure of GDP-bound dynamic microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Alushin, G.M, Lander, G.C, Kellogg, E.H, Zhang, R, Baker, D, Nogales, E. | | Deposit date: | 2014-02-19 | | Release date: | 2014-06-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | High-Resolution Microtubule Structures Reveal the Structural Transitions in alpha beta-Tubulin upon GTP Hydrolysis.
Cell(Cambridge,Mass.), 157, 2014
|
|
3JVE
 
 | |
1MOW
 
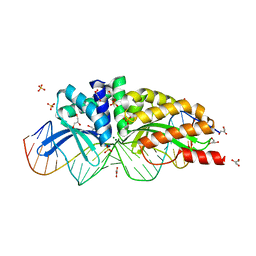 | | E-DreI | | Descriptor: | 5'-D(*CP*CP*AP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*AP*GP*TP*TP*CP*CP*GP*GP*CP*G)-3', 5'-D(*CP*GP*CP*CP*GP*GP*AP*AP*CP*TP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*TP*GP*G)-3', GLYCEROL, ... | | Authors: | Chevalier, B.S, Kortemme, T, Chadsey, M.S, Baker, D, Monnat Jr, R.J, Stoddard, B.L. | | Deposit date: | 2002-09-10 | | Release date: | 2002-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design, Activity and Structure of a Highly Specific Artificial Endonuclease
Mol.Cell, 10, 2002
|
|
1MPU
 
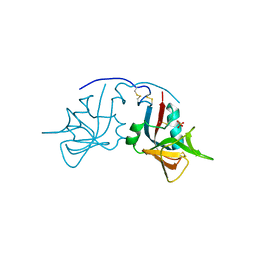 | | Crystal Structure of the free human NKG2D immunoreceptor | | Descriptor: | NKG2-D type II integral membrane protein, PHOSPHATE ION | | Authors: | McFarland, B.J, Kortemme, T, Baker, D, Strong, R.K. | | Deposit date: | 2002-09-12 | | Release date: | 2003-04-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Symmetry Recognizing Asymmetry: Analysis of the Interactions between the C-Type Lectin-like Immunoreceptor NKG2D and MHC
Class I-like Ligands
Structure, 11, 2003
|
|
3J6G
 
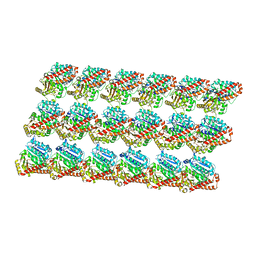 | | Minimized average structure of microtubules stabilized by taxol | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Alushin, G.M, Lander, G.C, Kellogg, E.H, Zhang, R, Baker, D, Nogales, E. | | Deposit date: | 2014-02-19 | | Release date: | 2014-06-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | High-Resolution Microtubule Structures Reveal the Structural Transitions in alpha beta-Tubulin upon GTP Hydrolysis.
Cell(Cambridge,Mass.), 157, 2014
|
|
5TVV
 
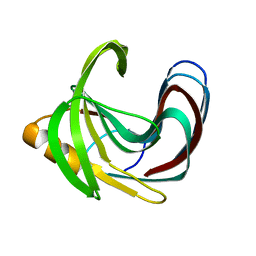 | | Computationally Designed Fentanyl Binder - Fen49* Apo | | Descriptor: | Endo-1,4-beta-xylanase A, POTASSIUM ION | | Authors: | Bick, M.J, Greisen, P.J, Morey, K.J, Antunes, A.S, La, D, Sankaran, B, Reymond, L, Johnsson, K, Medford, J.I, Baker, D. | | Deposit date: | 2016-11-10 | | Release date: | 2017-10-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Computational design of environmental sensors for the potent opioid fentanyl.
Elife, 6, 2017
|
|
2GIY
 
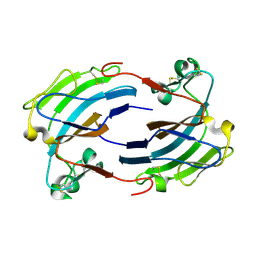 | |
3J9G
 
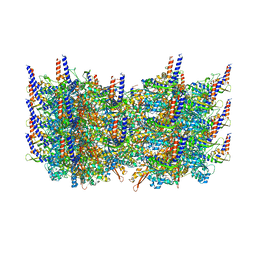 | | Atomic model of the VipA/VipB, the type six secretion system contractile sheath of Vibrio cholerae from cryo-EM | | Descriptor: | VipA, VipB | | Authors: | Kudryashev, M, Wang, R.Y.-R, Brackmann, M, Scherer, S, Maier, T, Baker, D, DiMaio, F, Stahlberg, H, Egelman, E.H, Basler, M. | | Deposit date: | 2015-01-16 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the Type VI Secretion System Contractile Sheath.
Cell(Cambridge,Mass.), 160, 2015
|
|
3J1V
 
 | |
4ILS
 
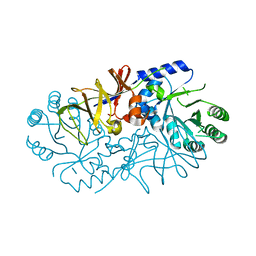 | | Crystal structure of engineered protein. northeast structural genomics Consortium target or117 | | Descriptor: | Engineered protein | | Authors: | Seetharaman, J, Lew, S, Nivon, L, Baker, D, Bjelic, S, Ciccosanti, C, Sahdev, S, Xiao, R, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-12-31 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of engineered protein. northeast structural genomics Consortium target or117
To be Published
|
|
2A3J
 
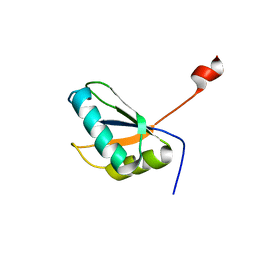 | | Structure of URNdesign, a complete computational redesign of human U1A protein | | Descriptor: | U1 small nuclear ribonucleoprotein A | | Authors: | Varani, G, Dobson, N, Dantas, G, Baker, D. | | Deposit date: | 2005-06-24 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Structural Validation of the Computational Redesign of Human U1A Protein
Structure, 14, 2006
|
|
4J29
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR258. | | Descriptor: | Engineered Protein OR258 | | Authors: | Vorobiev, S, Su, M, Koga, R, Seetharaman, J, Koga, N, Mao, L, Xiao, R, Kohan, E, Castelllanos, J, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-02-04 | | Release date: | 2013-03-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Engineered Protein OR258.
To be Published
|
|
4JVV
 
 | | Crystal structure of the evolved variant of the computationally designed serine hydrolase, OSH55.4_H1, covalently bound with diisopropyl fluorophosphate (DFP), Northeast Structural Genomics Consortium (NESG) Target OR273 | | Descriptor: | evolved variant of the computationally designed serine hydrolase | | Authors: | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Tong, S, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-03-26 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.288 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
