7ZWB
 
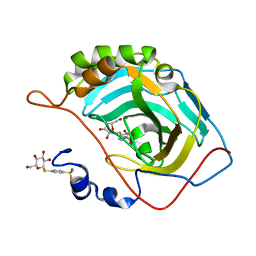 | | human Carbonic Anhydrase II in complex with 4-(((2S,3R,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl)thio)benzenesulfonamide | | Descriptor: | 4-[(2~{S},3~{R},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]sulfanylbenzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2022-05-19 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | First-in-Class Dual Targeting Compounds for the Management of Seizures in Glucose Transporter Type 1 Deficiency Syndrome.
J.Med.Chem., 66, 2023
|
|
6Y1Z
 
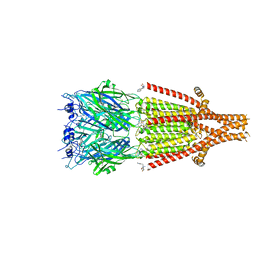 | | Mouse serotonin 5HT3 receptor in complex with palonosetron | | Descriptor: | (3~{a}~{S})-2-[(3~{S})-1-azabicyclo[2.2.2]octan-3-yl]-3~{a},4,5,6-tetrahydro-3~{H}-benzo[de]isoquinolin-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zarkadas, E, Perot, J, Nury, H. | | Deposit date: | 2020-02-14 | | Release date: | 2020-03-04 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | The Binding of Palonosetron and Other Antiemetic Drugs to the Serotonin 5-HT3 Receptor.
Structure, 28, 2020
|
|
5LG8
 
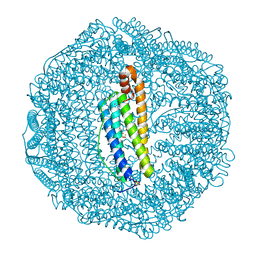 | | Human L-type ferritin iron loaded for 60 minutes | | Descriptor: | CADMIUM ION, FE (III) ION, Ferritin light chain, ... | | Authors: | Pozzi, C, Di Pisa, F, Mangani, S. | | Deposit date: | 2016-07-06 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Chemistry at the protein-mineral interface in L-ferritin assists the assembly of a functional ( mu (3)-oxo)Tris[( mu (2)-peroxo)] triiron(III) cluster.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5LG2
 
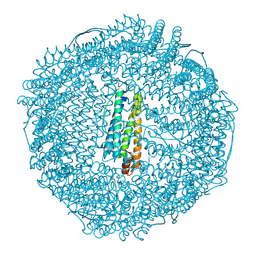 | | Horse L type ferritin iron loaded for 60 minutes | | Descriptor: | CADMIUM ION, CHLORIDE ION, FE (III) ION, ... | | Authors: | Pozzi, C, Di Pisa, F, Mangani, S. | | Deposit date: | 2016-07-05 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Chemistry at the protein-mineral interface in L-ferritin assists the assembly of a functional ( mu (3)-oxo)Tris[( mu (2)-peroxo)] triiron(III) cluster.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6Y9R
 
 | | Crystal structure of GSK-3b in complex with the 1H-indazole-3-carboxamide inhibitor 2 | | Descriptor: | ACETATE ION, GLYCEROL, Glycogen synthase kinase-3 beta, ... | | Authors: | Krapp, S, Griessner, A, Blaesse, M, Buonfiglio, R, Ombrato, R. | | Deposit date: | 2020-03-10 | | Release date: | 2020-05-20 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Discovery of Novel Imidazopyridine GSK-3 beta Inhibitors Supported by Computational Approaches.
Molecules, 25, 2020
|
|
2IVN
 
 | | Structure of UP1 protein | | Descriptor: | GLYCEROL, MAGNESIUM ION, O-SIALOGLYCOPROTEIN ENDOPEPTIDASE, ... | | Authors: | Hecker, A, Leulliot, N, Graille, M, Dorlet, P, Quevillon-Cheruel, S, Ulryck, N, Van Tilbeurgh, H, Forterre, P. | | Deposit date: | 2006-06-14 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | An Archaeal Orthologue of the Universal Protein Kae1 is an Iron Metalloprotein which Exhibits Atypical DNA-Binding Properties and Apurinic-Endonuclease Activity in Vitro.
Nucleic Acids Res., 35, 2007
|
|
3RKR
 
 | | Crystal structure of a metagenomic short-chain oxidoreductase (SDR) in complex with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, Short chain oxidoreductase | | Authors: | Mayerhofer, H, Bijtenhoorn, P, Streit, W.R, Mueller-Dieckmann, J. | | Deposit date: | 2011-04-18 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | A novel metagenomic short-chain dehydrogenase/reductase attenuates Pseudomonas aeruginosa biofilm formation and virulence on Caenorhabditis elegans.
Plos One, 6, 2011
|
|
2IVO
 
 | | Structure of UP1 protein | | Descriptor: | TUNGSTATE(VI)ION, UP1 | | Authors: | Hecker, A, Leulliot, N, Graille, M, Dorlet, P, Quevillon-Cheruel, S, Ulryck, N, Van Tilbeurgh, H, Forterre, P. | | Deposit date: | 2006-06-14 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | An Archaeal Orthologue of the Universal Protein Kae1 is an Iron Metalloprotein which Exhibits Atypical DNA-Binding Properties and Apurinic-Endonuclease Activity in Vitro.
Nucleic Acids Res., 35, 2007
|
|
2IVP
 
 | | Structure of UP1 protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FE (II) ION, O-SIALOGLYCOPROTEIN ENDOPEPTIDASE | | Authors: | Hecker, A, Leulliot, N, Graille, M, Dorlet, P, Quevillon-Cheruel, S, Ulryck, N, Van Tilbeurgh, H, Forterre, P. | | Deposit date: | 2006-06-14 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An Archaeal Orthologue of the Universal Protein Kae1 is an Iron Metalloprotein which Exhibits Atypical DNA-Binding Properties and Apurinic-Endonuclease Activity in Vitro.
Nucleic Acids Res., 35, 2007
|
|
3LK8
 
 | |
6HWN
 
 | | Structure of Thermus thermophilus ClpP in complex with a tripeptide. | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, DI(HYDROXYETHYL)ETHER, Unknown tripeptide | | Authors: | Felix, J, Schanda, P, Fraga, H, Morlot, C. | | Deposit date: | 2018-10-12 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mechanism of the allosteric activation of the ClpP protease machinery by substrates and active-site inhibitors.
Sci Adv, 5, 2019
|
|
3DSH
 
 | | Crystal structure of dimeric interferon regulatory factor 5 (IRF-5) transactivation domain | | Descriptor: | Interferon regulatory factor 5 | | Authors: | Chen, W, Lam, S.S, Srinath, H, Jiang, Z, Correia, J.J, Schiffer, C, Fitzgerald, K.A, Lin, K, Royer Jr, W.E. | | Deposit date: | 2008-07-12 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into interferon regulatory factor activation from the crystal structure of dimeric IRF5.
Nat.Struct.Mol.Biol., 15, 2008
|
|
8OKL
 
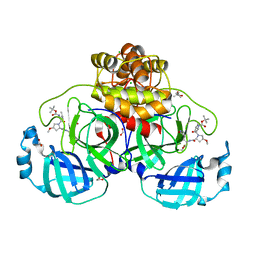 | | Crystal structure of F2F-2020185-01X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-03-28 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Broad-spectrum coronavirus 3C-like protease peptidomimetic inhibitors effectively block SARS-CoV-2 replication in cells: Design, synthesis, biological evaluation, and X-ray structure determination.
Eur.J.Med.Chem., 253, 2023
|
|
8OKK
 
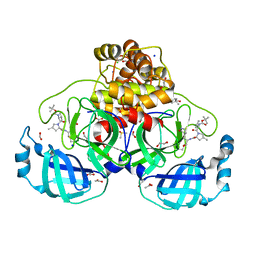 | | Crystal structure of F2F-2020184-00X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-03-28 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Broad-spectrum coronavirus 3C-like protease peptidomimetic inhibitors effectively block SARS-CoV-2 replication in cells: Design, synthesis, biological evaluation, and X-ray structure determination.
Eur.J.Med.Chem., 253, 2023
|
|
8OKM
 
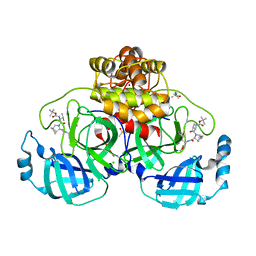 | | Crystal structure of F2F-2020197-00X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-03-28 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Broad-spectrum coronavirus 3C-like protease peptidomimetic inhibitors effectively block SARS-CoV-2 replication in cells: Design, synthesis, biological evaluation, and X-ray structure determination.
Eur.J.Med.Chem., 253, 2023
|
|
8OKN
 
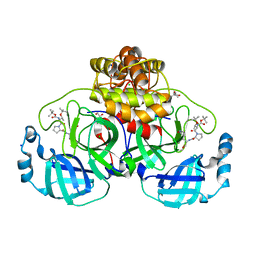 | | Crystal structure of F2F-2020198-00X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-03-28 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Broad-spectrum coronavirus 3C-like protease peptidomimetic inhibitors effectively block SARS-CoV-2 replication in cells: Design, synthesis, biological evaluation, and X-ray structure determination.
Eur.J.Med.Chem., 253, 2023
|
|
3EL9
 
 | | Crystal structure of atazanavir (ATV) in complex with a multidrug HIV-1 protease (V82T/I84V) | | Descriptor: | (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5, 6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER, ACETATE ION, ... | | Authors: | Prabu-Jeyabalan, M, King, N, Schiffer, C. | | Deposit date: | 2008-09-21 | | Release date: | 2009-09-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Extreme Entropy-Enthalpy Compensation in a Drug-Resistant Variant of HIV-1 Protease.
Acs Chem.Biol., 7, 2012
|
|
6HWM
 
 | | Structure of Thermus thermophilus ClpP in complex with bortezomib | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, DI(HYDROXYETHYL)ETHER, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE | | Authors: | Felix, J, Schanda, P, Fraga, H, Morlot, C. | | Deposit date: | 2018-10-12 | | Release date: | 2019-09-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism of the allosteric activation of the ClpP protease machinery by substrates and active-site inhibitors.
Sci Adv, 5, 2019
|
|
4OYN
 
 | | Fifteen minutes iron loaded human H ferritin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BICINE, CHLORIDE ION, ... | | Authors: | Pozzi, C, Di Pisa, F, Mangani, S. | | Deposit date: | 2014-02-12 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Iron binding to human heavy-chain ferritin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3EL5
 
 | |
3SOB
 
 | | The structure of the first YWTD beta propeller domain of LRP6 in complex with a FAB | | Descriptor: | CALCIUM ION, Low-density lipoprotein receptor-related protein 6, antibody heavy chain, ... | | Authors: | Wang, W, Bourhis, E, Tam, C, Zhang, Y, Rouge, L, Wu, Y, Franke, Y, Cochran, A.G. | | Deposit date: | 2011-06-30 | | Release date: | 2011-09-21 | | Last modified: | 2014-05-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Wnt antagonists bind through a short peptide to the first beta-propeller domain of LRP5/6.
Structure, 19, 2011
|
|
3EL4
 
 | | Crystal structure of inhibitor saquinavir (SQV) complexed with the multidrug HIV-1 protease variant L63P/V82T/I84V | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, ACETATE ION, Protease | | Authors: | Prabu-Jeyabalan, M, King, N, Schiffer, C. | | Deposit date: | 2008-09-19 | | Release date: | 2009-09-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Extreme Entropy-Enthalpy Compensation in a Drug-Resistant Variant of HIV-1 Protease.
Acs Chem.Biol., 7, 2012
|
|
3SOQ
 
 | | The structure of the first YWTD beta propeller domain of LRP6 in complex with a DKK1 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Dickkopf-related protein 1, ... | | Authors: | Wang, W, Bourhis, E, Zhang, Y, Rouge, L, Wu, Y, Franke, Y, Cochran, A.G. | | Deposit date: | 2011-06-30 | | Release date: | 2011-09-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Wnt antagonists bind through a short peptide to the first beta-propeller domain of LRP5/6.
Structure, 19, 2011
|
|
2R8J
 
 | | Structure of the Eukaryotic DNA Polymerase eta in complex with 1,2-d(GpG)-cisplatin containing DNA | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, Cisplatin, ... | | Authors: | Carell, T, Alt, A, Lammens, K. | | Deposit date: | 2007-09-11 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Bypass of DNA lesions generated during anticancer treatment with cisplatin by DNA polymerase eta
Science, 318, 2007
|
|
1N95
 
 | | Aryl Tetrahydrophyridine Inhbitors of Farnesyltranferase: Glycine, Phenylalanine and Histidine Derivatives | | Descriptor: | 1-[2-(4-CYANO-BENZYLAMINO)-3-(3-METHYL-3H-IMIDAZOL-4-YL)-PROPIONYL]-5-NAPHTHALEN-1-YL-1,2,3,6-TETRAHYDRO-PYRIDINE-4-CARBONITRILE, ALPHA-HYDROXYFARNESYLPHOSPHONIC ACID, Protein farnesyltransferase alpha subunit, ... | | Authors: | Gwaltney II, S.L, O'Conner, S.J, Nelson, L.T, Sullivan, G.M, Imade, H, Wang, W, Hasvold, L, Li, Q, Cohen, J, Gu, W.Z. | | Deposit date: | 2002-11-22 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Aryl tetrahydropyridine inhibitors of farnesyltransferase: glycine, phenylalanine and histidine derivatives.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
