9NBD
 
 | | AUGMIN Dimer | | Descriptor: | AUGMIN subunit 1, AUGMIN subunit 3, AUGMIN subunit 4, ... | | Authors: | Ashaduzzaman, M, Al-Bassam, J, Taheri, A. | | Deposit date: | 2025-02-13 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Cryo-EM structures of the Plant Augmin reveal its intertwined coiled-coil assembly, antiparallel dimerization and NEDD1 binding mechanisms.
Biorxiv, 2025
|
|
9NA9
 
 | | Augmin1345-Extended-Tripod | | Descriptor: | AUGMIN subunit 1, AUGMIN subunit 3, AUGMIN subunit 4, ... | | Authors: | Ashaduzzaman, M, Al-Bassam, J, Taheri, A. | | Deposit date: | 2025-02-11 | | Release date: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Cryo-EM structures of the Plant Augmin reveal its intertwined coiled-coil assembly, antiparallel dimerization and NEDD1 binding mechanisms.
Biorxiv, 2025
|
|
9NBI
 
 | | AUGMIN(V junction)/NEDD1(WD) | | Descriptor: | AUGMIN subunit 2, AUGMIN subunit 3, AUGMIN subunit 5, ... | | Authors: | Ashaduzzaman, M, Al-Bassam, J. | | Deposit date: | 2025-02-13 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Cryo-EM structures of the Plant Augmin reveal its intertwined coiled-coil assembly, antiparallel dimerization and NEDD1 binding mechanisms.
Biorxiv, 2025
|
|
9NBA
 
 | | Augmin/V junction(open) | | Descriptor: | AUGMIN subunit 2, AUGMIN subunit 3, AUGMIN subunit 5, ... | | Authors: | Ashaduzzaman, M, Al-Bassam, J, Taheri, A. | | Deposit date: | 2025-02-13 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Cryo-EM structures of the Plant Augmin reveal its intertwined coiled-coil assembly, antiparallel dimerization and NEDD1 binding mechanisms.
Biorxiv, 2025
|
|
9NA8
 
 | | Augmin1345 Extended-body | | Descriptor: | AUGMIN subunit 1, AUGMIN subunit 3, AUGMIN subunit 4, ... | | Authors: | Ashaduzzaman, M, Al-Bassam, J, Taheri, A. | | Deposit date: | 2025-02-11 | | Release date: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of the Plant Augmin reveal its intertwined coiled-coil assembly, antiparallel dimerization and NEDD1 binding mechanisms.
Biorxiv, 2025
|
|
6RRH
 
 | | GOLGI ALPHA-MANNOSIDASE II | | Descriptor: | 1,2-ETHANEDIOL, Alpha-mannosidase 2, ZINC ION | | Authors: | Armstrong, Z, Lahav, D, Johnson, R, Kuo, C.L, Beenakker, T.J.M, de Boer, C, Wong, C.S, van Rijssel, E.R, Debets, M, Geurink, P.P, Ovaa, H, van der Stelt, M, Codee, J.D.C, Aerts, J.M.F.G, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2019-05-18 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Manno- epi -cyclophellitols Enable Activity-Based Protein Profiling of Human alpha-Mannosidases and Discovery of New Golgi Mannosidase II Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
6RRJ
 
 | | GOLGI ALPHA-MANNOSIDASE II in complex with 5-(Adamantan-1yl-methoxy)-pentyl 2,5-dideoxy-2,5-imino-D-talo-hexonamide | | Descriptor: | (2~{S},3~{R},4~{S},5~{R})-~{N}-[5-(1-adamantylmethoxy)pentyl]-5-(hydroxymethyl)-3,4-bis(oxidanyl)pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, Alpha-mannosidase 2, ... | | Authors: | Armstrong, Z, Lahav, D, Johnson, R, Kuo, C.L, Beenakker, T.J.M, de Boer, C, Wong, C.S, van Rijssel, E.R, Debets, M, Geurink, P.P, Ovaa, H, van der Stelt, M, Codee, J.D.C, Aerts, J.M.F.G, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2019-05-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Manno- epi -cyclophellitols Enable Activity-Based Protein Profiling of Human alpha-Mannosidases and Discovery of New Golgi Mannosidase II Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
6RRU
 
 | | GOLGI ALPHA-MANNOSIDASE II in complex with (5R,6R,7S,8S)-5,6,7,8-tetrahydro-5-(hydroxymethyl)-3-(3-phenylpropyl)imidazo[1,2-a]pyridine-6,7,8-triol | | Descriptor: | (5~{R},6~{R},7~{S},8~{S})-5-(hydroxymethyl)-2-(3-phenylpropyl)-5,6,7,8-tetrahydroimidazo[1,2-a]pyridine-6,7,8-triol, Alpha-mannosidase 2, ZINC ION | | Authors: | Armstrong, Z, Lahav, D, Johnson, R, Kuo, C.L, Beenakker, T.J.M, de Boer, C, Wong, C.S, van Rijssel, E.R, Debets, M, Geurink, P.P, Ovaa, H, van der Stelt, M, Codee, J.D.C, Aerts, J.M.F.G, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2019-05-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Manno- epi -cyclophellitols Enable Activity-Based Protein Profiling of Human alpha-Mannosidases and Discovery of New Golgi Mannosidase II Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
6RRX
 
 | | GOLGI ALPHA-MANNOSIDASE II in complex with (2S,3R)-2-(Hydroxymethyl)-3-piperidinol | | Descriptor: | (2~{S},3~{R})-2-(hydroxymethyl)piperidin-3-ol, 1,2-ETHANEDIOL, Alpha-mannosidase 2, ... | | Authors: | Armstrong, Z, Lahav, D, Johnson, R, Kuo, C.L, Beenakker, T.J.M, de Boer, C, Wong, C.S, van Rijssel, E.R, Debets, M, Geurink, P.P, Ovaa, H, van der Stelt, M, Codee, J.D.C, Aerts, J.M.F.G, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2019-05-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Manno- epi -cyclophellitols Enable Activity-Based Protein Profiling of Human alpha-Mannosidases and Discovery of New Golgi Mannosidase II Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
5W5O
 
 | |
6RS0
 
 | | GOLGI ALPHA-MANNOSIDASE II in complex with (2S,3S,4R,5R)-1-(2-(Benzyloxy)ethyl)-2-(hydroxymethyl)piperidine-3,4,5-triol | | Descriptor: | (2~{S},3~{S},4~{R})-2-(hydroxymethyl)-1-(2-phenylmethoxyethyl)piperidine-3,4,5-triol, 1,2-ETHANEDIOL, Alpha-mannosidase 2, ... | | Authors: | Armstrong, Z, Lahav, D, Johnson, R, Kuo, C.L, Beenakker, T.J.M, de Boer, C, Wong, C.S, van Rijssel, E.R, Debets, M, Geurink, P.P, Ovaa, H, van der Stelt, M, Codee, J.D.C, Aerts, J.M.F.G, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2019-05-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Manno- epi -cyclophellitols Enable Activity-Based Protein Profiling of Human alpha-Mannosidases and Discovery of New Golgi Mannosidase II Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
6RRN
 
 | | GOLGI ALPHA-MANNOSIDASE II in complex with pentyl 2,5-dideoxy-2,5-imino-D-talo-hexonamide | | Descriptor: | (2~{S},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)-~{N}-pentyl-pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, Alpha-mannosidase 2, ... | | Authors: | Armstrong, Z, Lahav, D, Johnson, R, Kuo, C.L, Beenakker, T.J.M, de Boer, C, Wong, C.S, van Rijssel, E.R, Debets, M, Geurink, P.P, Ovaa, H, van der Stelt, M, Codee, J.D.C, Aerts, J.M.F.G, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2019-05-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Manno- epi -cyclophellitols Enable Activity-Based Protein Profiling of Human alpha-Mannosidases and Discovery of New Golgi Mannosidase II Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
6RRY
 
 | | GOLGI ALPHA-MANNOSIDASE II in complex with (2S,3R)-2-(Hydroxymethyl)-1,2,3,6-tetrahydro-3-pyridinol | | Descriptor: | (2~{S},3~{R})-2-(hydroxymethyl)-1,2,3,6-tetrahydropyridin-3-ol, 1,2-ETHANEDIOL, Alpha-mannosidase 2, ... | | Authors: | Armstrong, Z, Lahav, D, Johnson, R, Kuo, C.L, Beenakker, T.J.M, de Boer, C, Wong, C.S, van Rijssel, E.R, Debets, M, Geurink, P.P, Ovaa, H, van der Stelt, M, Codee, J.D.C, Aerts, J.M.F.G, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2019-05-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Manno- epi -cyclophellitols Enable Activity-Based Protein Profiling of Human alpha-Mannosidases and Discovery of New Golgi Mannosidase II Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
8WGW
 
 | | Local refinement of RBD-ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Wei, X, Zhang, Z. | | Deposit date: | 2023-09-22 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The receptor binding domain of SARS-CoV-2 Omicron subvariants targets Siglec-9 to decrease its immunogenicity by preventing macrophage phagocytosis.
Nat.Immunol., 25, 2024
|
|
8WGV
 
 | | BA.2(S375) Spike (S6P)/hACE2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Wei, X, Zhang, Z. | | Deposit date: | 2023-09-22 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | The receptor binding domain of SARS-CoV-2 Omicron subvariants targets Siglec-9 to decrease its immunogenicity by preventing macrophage phagocytosis.
Nat.Immunol., 25, 2024
|
|
5W5J
 
 | |
6RQZ
 
 | | GOLGI ALPHA-MANNOSIDASE II complex with Manno-epi-cyclophellitol aziridine | | Descriptor: | (1~{R},2~{S},3~{R},4~{R},5~{S},6~{R})-5-azanyl-6-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, Alpha-mannosidase 2, ... | | Authors: | Armstrong, Z, Lahav, D, Johnson, R, Kuo, C.L, Beenakker, T.J.M, de Boer, C, Wong, C.S, Debets, M, van der Stelt, M, Codee, J.D.C, Aerts, J.M.F.G, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2019-05-16 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Manno- epi -cyclophellitols Enable Activity-Based Protein Profiling of Human alpha-Mannosidases and Discovery of New Golgi Mannosidase II Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
6RRW
 
 | | GOLGI ALPHA-MANNOSIDASE II in complex with (2R,3R,4R,5S)-1-(5-{[4-(3,4-Dihydro-2H-1,5-benzodioxepin-7-yl)benzyl]oxy}pentyl)-2-(hydroxymethyl)-3,4,5-piperidinetriol | | Descriptor: | (2~{R},3~{R},4~{R},5~{S})-1-[5-[[4-(3,4-dihydro-2~{H}-1,5-benzodioxepin-7-yl)phenyl]methoxy]pentyl]-2-(hydroxymethyl)piperidine-3,4,5-triol, Alpha-mannosidase 2, ZINC ION | | Authors: | Armstrong, Z, Lahav, D, Johnson, R, Kuo, C.L, Beenakker, T.J.M, de Boer, C, Wong, C.S, van Rijssel, E.R, Debets, M, Geurink, P.P, Ovaa, H, van der Stelt, M, Codee, J.D.C, Aerts, J.M.F.G, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2019-05-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Manno- epi -cyclophellitols Enable Activity-Based Protein Profiling of Human alpha-Mannosidases and Discovery of New Golgi Mannosidase II Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
6RPC
 
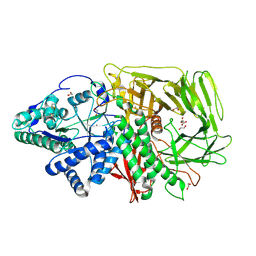 | | GOLGI ALPHA-MANNOSIDASE II | | Descriptor: | 1,2-ETHANEDIOL, Alpha-mannosidase 2, SUCCINIC ACID, ... | | Authors: | Armstrong, Z, Lahav, D, Johnson, R, Kuo, C.L, Beenakker, T.J.M, de Boer, C, Wong, C.S, Debets, M, van der Stelt, M, Codee, J.D.C, Aerts, J.M.F.G, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2019-05-14 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Manno- epi -cyclophellitols Enable Activity-Based Protein Profiling of Human alpha-Mannosidases and Discovery of New Golgi Mannosidase II Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
1HVC
 
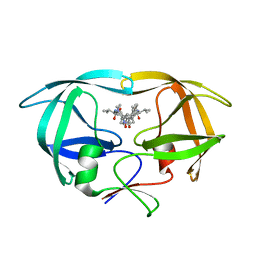 | | CRYSTAL STRUCTURE OF A TETHERED DIMER OF HIV-1 PROTEASE COMPLEXED WITH AN INHIBITOR | | Descriptor: | HIV-1 PROTEASE, N-{1-BENZYL-(2S,3S)-2,3-DIHYDROXY-4-[3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRYLAMINO]-5-PHENYL-PENTYL}-3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRAMIDE | | Authors: | Bhat, T.N, Baldwin, E.T, Erickson, J.W. | | Deposit date: | 1994-06-22 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a tethered dimer of HIV-1 proteinase complexed with an inhibitor.
Nat.Struct.Biol., 1, 1994
|
|
3NEW
 
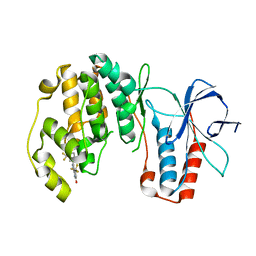 | | p38-alpha complexed with Compound 10 | | Descriptor: | 4-(trifluoromethyl)-3-[3-(trifluoromethyl)phenyl]-1,7-dihydro-6H-pyrazolo[3,4-b]pyridin-6-one, Mitogen-activated protein kinase 14 | | Authors: | Goedken, E.R, Comess, K.M, Sun, C, Argiriadi, M, Jia, Y, Quinn, C.M, Banach, D.L, Marcotte, D, Borhani, D. | | Deposit date: | 2010-06-09 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery and Characterization of Non-ATP Site Inhibitors of the Mitogen Activated Protein (MAP) Kinases.
Acs Chem.Biol., 6, 2011
|
|
3O17
 
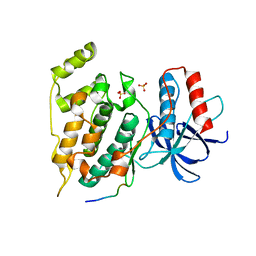 | | Crystal Structure of JNK1-alpha1 isoform | | Descriptor: | C-Jun-amino-terminal kinase-interacting protein 1, JIP1, 10MER PEPTIDE, ... | | Authors: | Abad-Zapatero, C. | | Deposit date: | 2010-07-20 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of JNK1-alpha1 isoform
TO BE PUBLISHED
|
|
1HPX
 
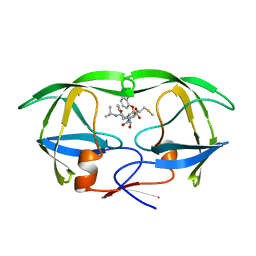 | | HIV-1 PROTEASE COMPLEXED WITH THE INHIBITOR KNI-272 | | Descriptor: | (4R)-N-tert-butyl-3-[(2S,3S)-2-hydroxy-3-({N-[(isoquinolin-5-yloxy)acetyl]-S-methyl-L-cysteinyl}amino)-4-phenylbutanoyl]-1,3-thiazolidine-4-carboxamide, HIV-1 PROTEASE | | Authors: | Bhat, T.N, Erickson, J.W. | | Deposit date: | 1995-05-18 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of HIV-1 protease with KNI-272, a tight-binding transition-state analog containing allophenylnorstatine.
Structure, 3, 1995
|
|
1ENH
 
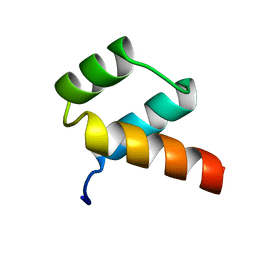 | | STRUCTURAL STUDIES OF THE ENGRAILED HOMEODOMAIN | | Descriptor: | ENGRAILED HOMEODOMAIN | | Authors: | Clarke, N.D, Kissinger, C.R, Desjarlais, J, Gilliland, G.L, Pabo, C.O. | | Deposit date: | 1994-05-20 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural studies of the engrailed homeodomain.
Protein Sci., 3, 1994
|
|
8HZ5
 
 | |
