4JOA
 
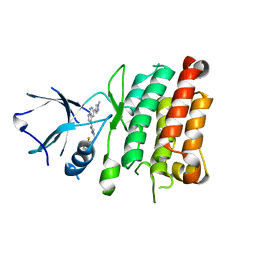 | | Crystal Structure of Human Anaplastic Lymphoma Kinase in complex with 7-azaindole based inhibitor | | Descriptor: | 3-[1-(2,5-difluorobenzyl)-1H-pyrazol-4-yl]-5-(1-methyl-1H-pyrazol-4-yl)-1H-pyrrolo[2,3-b]pyridine, ALK tyrosine kinase receptor | | Authors: | Hosahalli, S, Krishnamurthy, N.R, Lakshminarasimhan, A. | | Deposit date: | 2013-03-18 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of 7-azaindole based anaplastic lymphoma kinase (ALK) inhibitors: wild type and mutant (L1196M) active compounds with unique binding mode
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4JF5
 
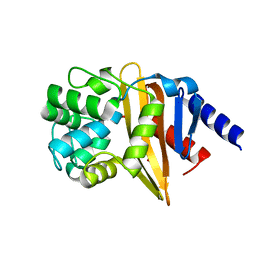 | | Structure of OXA-23 at pH 4.1 | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CITRATE ANION | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2013-02-27 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural Basis for Carbapenemase Activity of the OXA-23 beta-Lactamase from Acinetobacter baumannii.
Chem.Biol., 20, 2013
|
|
1SO8
 
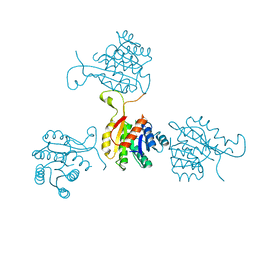 | | Abeta-bound human ABAD structure [also known as 3-hydroxyacyl-CoA dehydrogenase type II (Type II HADH), Endoplasmic reticulum-associated amyloid beta-peptide binding protein (ERAB)] | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase type II, CHLORIDE ION, SODIUM ION | | Authors: | Lustbader, J.W, Cirilli, M, Wu, H. | | Deposit date: | 2004-03-13 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ABAD directly links Abeta to mitochondrial toxicity in Alzheimer's disease.
Science, 304, 2004
|
|
4BL2
 
 | |
3VFF
 
 | |
4CPK
 
 | | Crystal structure of PBP2a double clinical mutant N146K-E150K from MRSA | | Descriptor: | CADMIUM ION, CHLORIDE ION, Penicillin binding protein 2 prime, ... | | Authors: | Otero, L.H, Rojas-Altuve, A, Hermoso, J.A. | | Deposit date: | 2014-02-07 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Disruption of Allosteric Response as an Unprecedented Mechanism of Resistance to Antibiotics.
J.Am.Chem.Soc., 136, 2014
|
|
4BL3
 
 | | Crystal structure of PBP2a clinical mutant N146K from MRSA | | Descriptor: | CADMIUM ION, CHLORIDE ION, PENICILLIN BINDING PROTEIN 2 PRIME, ... | | Authors: | Otero, L.H, Rojas-Altuve, A, Carrasco-Lopez, C, Hermoso, J.A. | | Deposit date: | 2013-04-30 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Disruption of Allosteric Response as an Unprecedented Mechanism of Resistance to Antibiotics.
J.Am.Chem.Soc., 136, 2014
|
|
3VFH
 
 | |
4NKN
 
 | |
4O97
 
 | | Crystal structure of matriptase in complex with inhibitor | | Descriptor: | N-(trans-4-aminocyclohexyl)-3,5-bis[(3-carbamimidoylbenzyl)oxy]benzamide, Peptide CGLR, Suppressor of tumorigenicity 14 protein | | Authors: | Rao, K.N, Chandra, B.R, Ashok, K.N, Chakshusmathi, G, Ramesh, K.S, Subramanya, H.S. | | Deposit date: | 2014-01-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-guided discovery of 1,3,5 tri-substituted benzenes as potent and selective matriptase inhibitors exhibiting in vivo antitumor efficacy.
Bioorg.Med.Chem., 22, 2014
|
|
4O9V
 
 | | Crystal structure of matriptase in complex with inhibitor | | Descriptor: | N-(trans-4-aminocyclohexyl)-3,5-bis(4-carbamimidoylphenoxy)benzamide, Peptide CGLR, Suppressor of tumorigenicity 14 protein | | Authors: | Rao, K.N, Chandra, B.R, Ashok, K.N, Chakshusmathi, G, Ramesh, K.S, Subramanya, H.S. | | Deposit date: | 2014-01-03 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided discovery of 1,3,5 tri-substituted benzenes as potent and selective matriptase inhibitors exhibiting in vivo antitumor efficacy.
Bioorg.Med.Chem., 22, 2014
|
|
4OE9
 
 | |
7JTZ
 
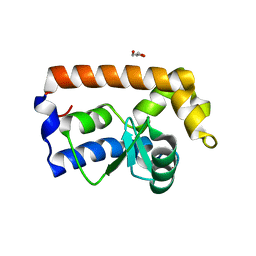 | | Yeast Glo3 GAP domain | | Descriptor: | ADP-ribosylation factor GTPase-activating protein GLO3, GLYCEROL, ZINC ION | | Authors: | Xie, B, Jackson, L.P. | | Deposit date: | 2020-08-18 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The Glo3 GAP crystal structure supports the molecular niche model for ArfGAPs in COPI coats.
Adv Biol Regul, 79, 2021
|
|
3K35
 
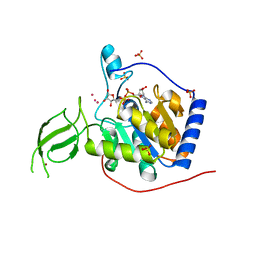 | | Crystal Structure of Human SIRT6 | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, NAD-dependent deacetylase sirtuin-6, SULFATE ION, ... | | Authors: | Pan, P.W, Dong, A, Qiu, W, Loppnau, P, Wang, J, Ravichandran, M, Bochkarev, A, Bountra, C, Weigelt, J, Arrowsmith, C.H, Min, J, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-01 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and biochemical functions of SIRT6.
J.Biol.Chem., 286, 2011
|
|
2OOZ
 
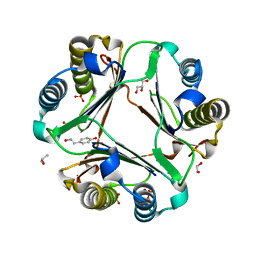 | | Macrophage Migration Inhibitory Factor (MIF) Complexed with OXIM6 (an OXIM Derivative Not Containing a Ring in its R-group) | | Descriptor: | 4-HYDROXYBENZALDEHYDE O-(3,3-DIMETHYLBUTANOYL)OXIME, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Crichlow, G.V, Al-Abed, Y, Lolis, E. | | Deposit date: | 2007-01-26 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Alternative chemical modifications reverse the binding orientation of a pharmacophore scaffold in the active site of macrophage migration inhibitory factor.
J.Biol.Chem., 282, 2007
|
|
2OOH
 
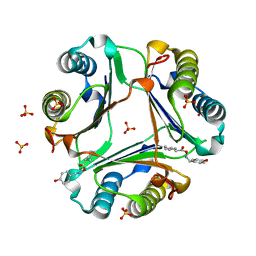 | | Crystal Structure of MIF bound to a Novel Inhibitor, OXIM-11 | | Descriptor: | 4-HYDROXYBENZALDEHYDE O-(CYCLOHEXYLCARBONYL)OXIME, GLYCEROL, Macrophage migration inhibitory factor, ... | | Authors: | Crichlow, G.V, Al-Abed, Y, Lolis, E. | | Deposit date: | 2007-01-25 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Alternative chemical modifications reverse the binding orientation of a pharmacophore scaffold in the active site of macrophage migration inhibitory factor.
J.Biol.Chem., 282, 2007
|
|
3PKI
 
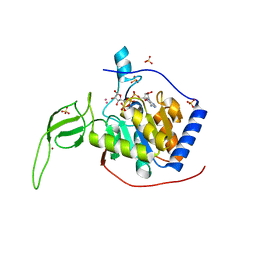 | | Human SIRT6 crystal structure in complex with ADP ribose | | Descriptor: | NAD-dependent deacetylase sirtuin-6, SULFATE ION, UNKNOWN ATOM OR ION, ... | | Authors: | Pan, P.W, Dong, A, Qiu, W, Loppnau, P, Wang, J, Ravichandran, M, Bochkarev, A, Bountra, C, Weigelt, J, Arrowsmith, C.H, Min, J, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-11 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure and biochemical functions of SIRT6.
J.Biol.Chem., 286, 2011
|
|
2OOW
 
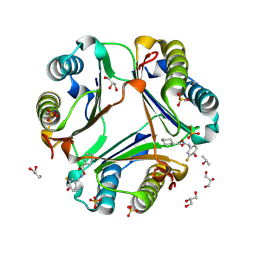 | | MIF Bound to a Fluorinated OXIM Derivative | | Descriptor: | 3-FLUORO-4-HYDROXYBENZALDEHYDE O-(CYCLOHEXYLCARBONYL)OXIME, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Crichlow, G.V, Al-Abed, Y, Lolis, E. | | Deposit date: | 2007-01-26 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Alternative chemical modifications reverse the binding orientation of a pharmacophore scaffold in the active site of macrophage migration inhibitory factor.
J.Biol.Chem., 282, 2007
|
|
6BTZ
 
 | | Crystal structure of the PI3KC2alpha C2 domain in space group C121 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, GLYCEROL, Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha, ... | | Authors: | Chen, K.-E, Collins, B.M. | | Deposit date: | 2017-12-08 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular Basis for Membrane Recruitment by the PX and C2 Domains of Class II Phosphoinositide 3-Kinase-C2 alpha.
Structure, 26, 2018
|
|
6BTY
 
 | | Crystal structure of the PI3KC2alpha C2 domain in space group P41212 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha | | Authors: | Chen, K.-E, Collins, B.M. | | Deposit date: | 2017-12-08 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.678 Å) | | Cite: | Molecular Basis for Membrane Recruitment by the PX and C2 Domains of Class II Phosphoinositide 3-Kinase-C2 alpha.
Structure, 26, 2018
|
|
3PKJ
 
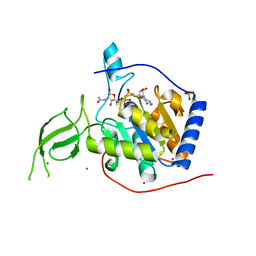 | | Human SIRT6 crystal structure in complex with 2'-N-Acetyl ADP ribose | | Descriptor: | NAD-dependent deacetylase sirtuin-6, SULFATE ION, UNKNOWN ATOM OR ION, ... | | Authors: | Pan, P.W, Dong, A, Qiu, W, Loppnau, P, Wang, J, Ravichandran, M, Walker, J.R, Bountra, C, Weigelt, J, Arrowsmith, C.H, Min, J, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-11 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure and biochemical functions of SIRT6.
J.Biol.Chem., 286, 2011
|
|
3PNW
 
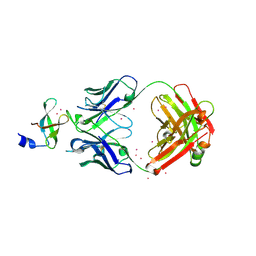 | | Crystal Structure of the tudor domain of human TDRD3 in complex with an anti-TDRD3 FAB | | Descriptor: | FAB heavy chain, FAB light chain, Tudor domain-containing protein 3, ... | | Authors: | Loppnau, P, Tempel, W, Wernimont, A.K, Lam, R, Ravichandran, M, Adams-Cioaba, M.A, Persson, H, Sidhu, S.S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Cossar, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-19 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | CDR-H3 Diversity Is Not Required for Antigen Recognition by Synthetic Antibodies.
J.Mol.Biol., 425, 2013
|
|
4JZI
 
 | | Crystal Structure of Matriptase in complex with Inhibitor". | | Descriptor: | N-(trans-4-aminocyclohexyl)-2,6-bis(4-carbamimidoylphenoxy)pyridine-4-carboxamide, Suppressor of tumorigenicity 14 protein | | Authors: | Subramanya, H.S, Chandra, R.B, Ashok, K.N, Chakshusmathi, G, Ramesh, K.S. | | Deposit date: | 2013-04-03 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Pyridyl Bis(oxy)dibenzimidamide Derivatives as Selective Matriptase Inhibitors
ACS MED.CHEM.LETT., 4, 2013
|
|
6BU0
 
 | | Crystal structure of the PI3KC2alpha C2 domain in complex with IP6 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, FORMIC ACID, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Chen, K.-E, Collins, B.M. | | Deposit date: | 2017-12-08 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.427 Å) | | Cite: | Molecular Basis for Membrane Recruitment by the PX and C2 Domains of Class II Phosphoinositide 3-Kinase-C2 alpha.
Structure, 26, 2018
|
|
6BUB
 
 | | Crystal structure of the PI3KC2alpha PX domain in space group P432 | | Descriptor: | GLYCEROL, Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha, SULFATE ION | | Authors: | Chen, K.-E, Collins, B.M. | | Deposit date: | 2017-12-09 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Molecular Basis for Membrane Recruitment by the PX and C2 Domains of Class II Phosphoinositide 3-Kinase-C2 alpha.
Structure, 26, 2018
|
|
