8WMJ
 
 | | structure of PSI-11CAC complex at Logrithmic growth phase | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-3,5,7,9,11,13,15-heptaen-1,17-diynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-(2,6,6-trimethylcyclohexen-1-yl)octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Zhang, S.M, Si, L, Li, M. | | Deposit date: | 2023-10-03 | | Release date: | 2024-05-29 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Growth phase-dependent reorganization of cryptophyte photosystem I antennae.
Commun Biol, 7, 2024
|
|
8WM6
 
 | | The structure of PSI-CAC(L-14)of R.salina at 2.7 angstroms resolution | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-3,5,7,9,11,13,15-heptaen-1,17-diynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-(2,6,6-trimethylcyclohexen-1-yl)octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Zhang, S.M, Si, L, Li, M. | | Deposit date: | 2023-10-03 | | Release date: | 2024-05-29 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Growth phase-dependent reorganization of cryptophyte photosystem I antennae.
Commun Biol, 7, 2024
|
|
8WNW
 
 | | the structure of PsaQ | | Descriptor: | 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, CHLOROPHYLL A, PsaQ | | Authors: | Zhang, S.M, Si, L, Li, M. | | Deposit date: | 2023-10-06 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Growth phase-dependent reorganization of cryptophyte photosystem I antennae.
Commun Biol, 7, 2024
|
|
9J3M
 
 | | ADP/Pi bound Arabidopsis ATP/ADP translocator AtNTT1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADP,ATP carrier protein 1, chloroplastic, ... | | Authors: | Lin, H.J, Huang, J, Li, T.M, Li, W.J, Su, N.N, Zhang, J.R, Wu, X.D, Fan, M.R. | | Deposit date: | 2024-08-08 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structure and mechanism of the plastid/parasite ATP/ADP translocator.
Nature, 641, 2025
|
|
9J3L
 
 | | ATP bound Arabidopsis ATP/ADP translocator AtNTT1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ADP,ATP carrier protein 1, chloroplastic, ... | | Authors: | Lin, H.J, Huang, J, Li, T.M, Li, W.J, Su, N.N, Zhang, J.R, Wu, X.D, Fan, M.R. | | Deposit date: | 2024-08-08 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structure and mechanism of the plastid/parasite ATP/ADP translocator.
Nature, 641, 2025
|
|
9J3N
 
 | | ATP bound Chlamydia pneumoniae ATP/ADP translocator NTT1(Inward open state) | | Descriptor: | 1D10, ADENOSINE-5'-TRIPHOSPHATE, ADP,ATP carrier protein 1 | | Authors: | Lin, H.J, Huang, J, Li, T.M, Li, W.J, Su, N.N, Zhang, J.R, Wu, X.D, Fan, M.R. | | Deposit date: | 2024-08-08 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structure and mechanism of the plastid/parasite ATP/ADP translocator.
Nature, 641, 2025
|
|
9J3O
 
 | | Chlamydia pneumoniae ATP/ADP translocator NTT1(Outward open state) | | Descriptor: | 1D10, ADP,ATP carrier protein 1 | | Authors: | Lin, H.J, Huang, J, Li, T.M, Li, W.J, Su, N.N, Zhang, J.R, Wu, X.D, Fan, M.R. | | Deposit date: | 2024-08-08 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and mechanism of the plastid/parasite ATP/ADP translocator.
Nature, 641, 2025
|
|
9J3J
 
 | | Arabidopsis ATP/ADP translocator AtNTT1 | | Descriptor: | ADP,ATP carrier protein 1, chloroplastic, nanobody: B-C8 | | Authors: | Lin, H.J, Huang, J, Li, T.M, Li, W.J, Su, N.N, Zhang, J.R, Wu, X.D, Fan, M.R. | | Deposit date: | 2024-08-08 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structure and mechanism of the plastid/parasite ATP/ADP translocator.
Nature, 641, 2025
|
|
4JV9
 
 | | Co-crystal structure of MDM2 with inhibitor (2S,5R,6S)-2-benzyl-5,6-bis(4-chlorophenyl)-4-methylmorpholin-3-one | | Descriptor: | (2S,5R,6S)-2-benzyl-5,6-bis(4-chlorophenyl)-4-methylmorpholin-3-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
4JWR
 
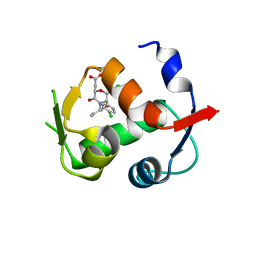 | |
9IK0
 
 | | Crystal structure of PrfaH encoded by IncX3 plasmids | | Descriptor: | GLYCEROL, MAGNESIUM ION, Transcription antitermination protein RfaH | | Authors: | Yang, J, Lu, Y, Yu, J, Cai, X, Wang, C, Lv, L, Liu, J. | | Deposit date: | 2024-06-26 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Comprehensive analysis of Enterobacteriaceae IncX plasmids reveals robust conjugation regulators PrfaH, H-NS, and conjugation-fitness tradeoff.
Commun Biol, 8, 2025
|
|
5DEY
 
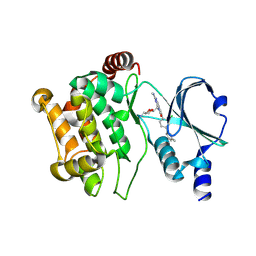 | | Crystal structure of PAK1 in complex with an inhibitor compound G-5555 | | Descriptor: | 8-[(trans-5-amino-1,3-dioxan-2-yl)methyl]-6-[2-chloro-4-(6-methylpyridin-2-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7(8H)-one, Serine/threonine-protein kinase PAK 1 | | Authors: | Oh, A, Tam, C, Wang, W. | | Deposit date: | 2015-08-26 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design of Selective PAK1 Inhibitor G-5555: Improving Properties by Employing an Unorthodox Low-pK a Polar Moiety.
Acs Med.Chem.Lett., 6, 2015
|
|
5DFP
 
 | | Crystal structure of PAK1 in complex with an inhibitor compound FRAX1036 | | Descriptor: | 6-[2-chloro-4-(6-methylpyrazin-2-yl)phenyl]-8-ethyl-2-{[2-(1-methylpiperidin-4-yl)ethyl]amino}pyrido[2,3-d]pyrimidin-7(8H)-one, DIMETHYL SULFOXIDE, Serine/threonine-protein kinase PAK 1 | | Authors: | Maksimoska, J, Marmorstein, R, Wang, W. | | Deposit date: | 2015-08-27 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design of Selective PAK1 Inhibitor G-5555: Improving Properties by Employing an Unorthodox Low-pK a Polar Moiety.
Acs Med.Chem.Lett., 6, 2015
|
|
5XZ3
 
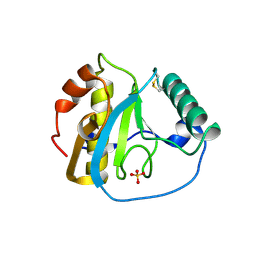 | | The X-ray structure of Apis mellifera PGRP-SA | | Descriptor: | Peptidoglycan-recognition protein, SULFATE ION | | Authors: | Liu, Y.J, Huang, J.X, Zhao, X.M, An, J.D. | | Deposit date: | 2017-07-11 | | Release date: | 2018-07-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of peptidoglycan recognition protein SA in Apis mellifera (Hymenoptera: Apidae).
Protein Sci., 27, 2018
|
|
2M6X
 
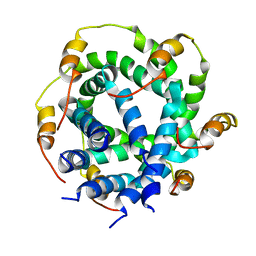 | |
1ZA7
 
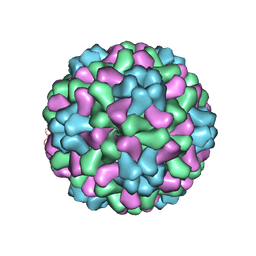 | | The crystal structure of salt stable cowpea cholorotic mottle virus at 2.7 angstroms resolution. | | Descriptor: | Coat protein | | Authors: | Bothner, B, Speir, J.A, Qu, C, Willits, D.A, Young, M.J, Johnson, J.E. | | Deposit date: | 2005-04-05 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Enhanced local symmetry interactions globally stabilize a mutant virus capsid that maintains infectivity and capsid dynamics.
J.Virol., 80, 2006
|
|
8WY4
 
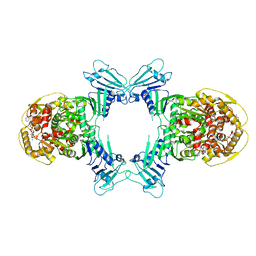 | | GajA tetramer with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Endonuclease GajA | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-10-30 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
8X5I
 
 | | Structure of ATP/Mg2+ bound Gabija GajA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Endonuclease GajA, MAGNESIUM ION | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-11-17 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
8WY5
 
 | | Structure of Gabija GajA in complex with DNA | | Descriptor: | CALCIUM ION, DNA (5'-D(P*AP*AP*AP*AP*TP*AP*AP*CP*CP*GP*GP*GP*TP*TP*AP*TP*TP*AP*A)-3'), DNA (5'-D(P*TP*TP*AP*AP*TP*AP*AP*CP*CP*CP*GP*GP*TP*TP*AP*TP*TP*TP*T)-3'), ... | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-10-30 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
8X5N
 
 | | Structure of ATP/Mg2+ bound Gabija GajA-GajB 4:4 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Endonuclease GajA, Gabija protein GajB, ... | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-11-17 | | Release date: | 2024-02-28 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
8X51
 
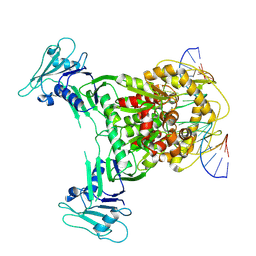 | | Structure of DNA-bound GajA dimer (focused refinement) | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*AP*AP*AP*AP*TP*AP*AP*CP*CP*GP*GP*GP*TP*TP*AP*TP*TP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*AP*TP*AP*AP*CP*CP*CP*GP*GP*TP*TP*AP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-11-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
2N2N
 
 | |
2N31
 
 | |
8DVG
 
 | | Structure of KRAS WT(7-16)-HLA-A*03:01 | | Descriptor: | Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, HLA class I histocompatibility antigen, ... | | Authors: | Wright, K.M, Miller, M, Gabelli, S.B. | | Deposit date: | 2022-07-28 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Hydrophobic interactions dominate the recognition of a KRAS G12V neoantigen.
Nat Commun, 14, 2023
|
|
6A0Q
 
 | | The crystal structure of Lpg2622_E64 complex | | Descriptor: | Lpg2622, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE | | Authors: | Gong, X, Ge, H. | | Deposit date: | 2018-06-06 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of the hypothetical protein Lpg2622, a new member of the C1 family peptidases from Legionella pneumophila
FEBS Lett., 592, 2018
|
|
