6Z10
 
 | | Crystal structure of a humanized (K18E, K269N) rat succinate receptor SUCNR1 (GPR91) in complex with a nanobody and antagonist | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-[2-[[3-[4-chloranyl-2-fluoranyl-5-[(3~{R})-piperidin-3-yl]oxy-phenyl]-2-fluoranyl-phenyl]carbonylamino]-5-fluoranyl-phenyl]ethanoic acid, CHLORIDE ION, ... | | Authors: | Haffke, M, Villard, F. | | Deposit date: | 2020-05-11 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.269 Å) | | Cite: | Discovery and Optimization of Novel SUCNR1 Inhibitors: Design of Zwitterionic Derivatives with a Salt Bridge for the Improvement of Oral Exposure.
J.Med.Chem., 63, 2020
|
|
3FBX
 
 | | Crystal structure of the lysosomal 66.3 kDa protein from mouse solved by S-SAD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Lakomek, K, Dickmanns, A, Mueller, U, Ficner, R. | | Deposit date: | 2008-11-20 | | Release date: | 2009-03-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | De novo sulfur SAD phasing of the lysosomal 66.3 kDa protein from mouse
Acta Crystallogr.,Sect.D, 65, 2009
|
|
5BNJ
 
 | | CDK8/CYCC IN COMPLEX WITH 8-{3-Chloro-5-[4-(1-methyl-1H-pyrazol-4-yl)-phenyl]-pyridin- 4-yl}-2,8-diaza-spiro[4.5]decan-1-one | | Descriptor: | 8-{3-chloro-5-[4-(1-methyl-1H-pyrazol-4-yl)phenyl]pyridin-4-yl}-2,8-diazaspiro[4.5]decan-1-one, Cyclin-C, Cyclin-dependent kinase 8, ... | | Authors: | Musil, D, Blagg, J, Wienke, D. | | Deposit date: | 2015-05-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | A selective chemical probe for exploring the role of CDK8 and CDK19 in human disease.
Nat.Chem.Biol., 11, 2015
|
|
6Q7A
 
 | |
6FZU
 
 | |
6Q6O
 
 | |
6Q7H
 
 | |
6G07
 
 | | RORGT (264-518;C455S) IN COMPLEX WITH INVERSE AGONIST "CPD-9" AND RIP140 PEPTIDE AT 1.66A | | Descriptor: | Nuclear receptor ROR-gamma, Nuclear receptor-interacting protein 1, ~{N}-[5-chloranyl-6-[(1~{S})-1-phenylethoxy]pyridin-3-yl]-2-(4-ethylsulfonylphenyl)ethanamide | | Authors: | Kallen, J. | | Deposit date: | 2018-03-16 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Optimizing a Weakly Binding Fragment into a Potent ROR gamma t Inverse Agonist with Efficacy in an in Vivo Inflammation Model.
J. Med. Chem., 61, 2018
|
|
6Q6M
 
 | |
6G05
 
 | | RORGT (264-518;C455S) IN COMPLEX WITH INVERSE AGONIST "CPD-2" AND RIP140 PEPTIDE AT 1.90A | | Descriptor: | 2-(4-ethylsulfonylphenyl)-~{N}-[4-phenyl-5-(phenylcarbonyl)-1,3-thiazol-2-yl]ethanamide, Nuclear receptor ROR-gamma, Nuclear receptor-interacting protein 1 | | Authors: | Kallen, J. | | Deposit date: | 2018-03-16 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimizing a Weakly Binding Fragment into a Potent ROR gamma t Inverse Agonist with Efficacy in an in Vivo Inflammation Model.
J. Med. Chem., 61, 2018
|
|
5N12
 
 | | Crystal structure of TCE treated rPPEP-1 | | Descriptor: | 2,2,2-tris-chloroethanol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Pro-Pro endopeptidase, ... | | Authors: | Pichlo, C, Schacherl, M, Baumann, U. | | Deposit date: | 2017-02-04 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Improved protein-crystal identification by using 2,2,2-trichloroethanol as a fluorescence enhancer.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
7LG6
 
 | | BG505 SOSIP.v5.2 in complex with VRC40.01 and RM19R Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Cottrell, C.A, Torres, J.L, Wu, N.R, Ward, A.B. | | Deposit date: | 2021-01-19 | | Release date: | 2021-09-15 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural basis of glycan276-dependent recognition by HIV-1 broadly neutralizing antibodies.
Cell Rep, 37, 2021
|
|
1YNA
 
 | | ENDO-1,4-BETA-XYLANASE, ROOM TEMPERATURE, PH 4.0 | | Descriptor: | ENDO-1,4-BETA-XYLANASE | | Authors: | Gruber, K, Kratky, C. | | Deposit date: | 1996-08-22 | | Release date: | 1997-02-12 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Thermophilic xylanase from Thermomyces lanuginosus: high-resolution X-ray structure and modeling studies.
Biochemistry, 37, 1998
|
|
6ZHY
 
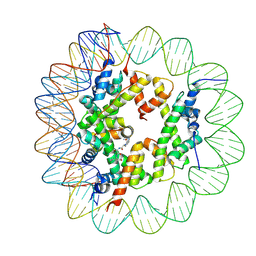 | | Cryo-EM structure of the regulatory linker of ALC1 bound to the nucleosome's acidic patch: hexasome class. | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1-like, DNA (110-MER) Widom 601 sequence, Histone H2A type 1, ... | | Authors: | Bacic, L, Gaullier, G, Deindl, S. | | Deposit date: | 2020-06-24 | | Release date: | 2020-12-23 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanistic Insights into Regulation of the ALC1 Remodeler by the Nucleosome Acidic Patch.
Cell Rep, 33, 2020
|
|
6ZHX
 
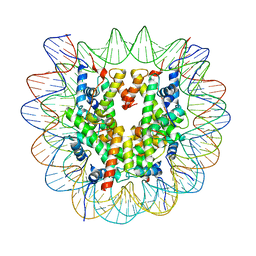 | | Cryo-EM structure of the regulatory linker of ALC1 bound to the nucleosome's acidic patch: nucleosome class. | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1-like, DNA (145-MER) Widom 601 sequence, Histone H2A type 1, ... | | Authors: | Bacic, L, Gaullier, G, Croll, T.I, Deindl, S. | | Deposit date: | 2020-06-24 | | Release date: | 2020-12-23 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanistic Insights into Regulation of the ALC1 Remodeler by the Nucleosome Acidic Patch.
Cell Rep, 33, 2020
|
|
1KNU
 
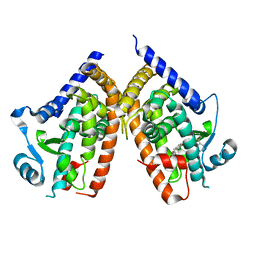 | | LIGAND BINDING DOMAIN OF THE HUMAN PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR GAMMA IN COMPLEX WITH A SYNTHETIC AGONIST | | Descriptor: | (S)-3-(4-(2-CARBAZOL-9-YL-ETHOXY)-PHENYL)-2-ETHOXY-PROPIONIC ACID, PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR GAMMA | | Authors: | Svensson, L.A, Mortensen, S.B, Fleckner, J, Woeldike, H.F. | | Deposit date: | 2001-12-19 | | Release date: | 2002-12-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel tricyclic-alpha-alkyloxyphenylpropionic acids: dual PPARalpha/gamma agonists with hypolipidemic and antidiabetic activity
J.MED.CHEM., 45, 2002
|
|
3RBX
 
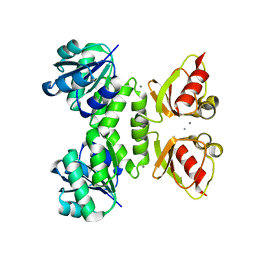 | | MthK RCK domain D184N mutant, Ca2+-bound | | Descriptor: | CALCIUM ION, Calcium-gated potassium channel mthK | | Authors: | Samakai, E, Rothberg, B.S. | | Deposit date: | 2011-03-30 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of multiple Ca2+-binding sites in a K+ channel RCK domain
Proc.Natl.Acad.Sci.USA, 2011
|
|
3RBZ
 
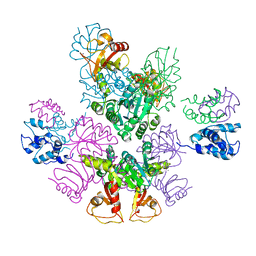 | | MthK channel, Ca2+-bound | | Descriptor: | CALCIUM ION, Calcium-gated potassium channel mthK | | Authors: | Taylor, A.B, Parfenova, L.V, Rothberg, B.S. | | Deposit date: | 2011-03-30 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structures of multiple Ca2+-binding sites in a K+ channel RCK domain
Proc.Natl.Acad.Sci.USA, 2011
|
|
6GX3
 
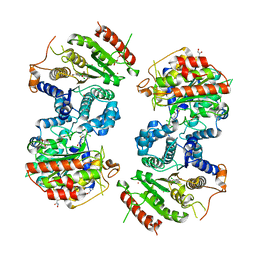 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with an hydroxamate 1 | | Descriptor: | 4-chloranyl-~{N}-oxidanyl-1-benzothiophene-2-carboxamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-06-26 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synthesis, Crystallization Studies, and in vitro Characterization of Cinnamic Acid Derivatives as SmHDAC8 Inhibitors for the Treatment of Schistosomiasis.
ChemMedChem, 13, 2018
|
|
6GXU
 
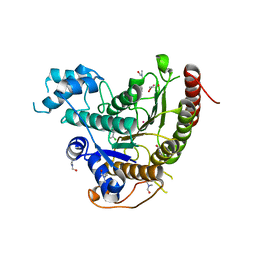 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with an hydroxamate 3 | | Descriptor: | (~{E})-3-[2-(4-chlorophenyl)sulfanylphenyl]-~{N}-oxidanyl-prop-2-enamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-06-27 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.917 Å) | | Cite: | Synthesis, Crystallization Studies, and in vitro Characterization of Cinnamic Acid Derivatives as SmHDAC8 Inhibitors for the Treatment of Schistosomiasis.
ChemMedChem, 13, 2018
|
|
6GXW
 
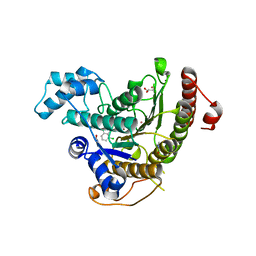 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with an hydroxamate 4 | | Descriptor: | (~{E})-3-[2-[[2,6-bis(chloranyl)phenyl]methoxy]phenyl]-~{N}-oxidanyl-prop-2-enamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-06-27 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.071 Å) | | Cite: | Synthesis, Crystallization Studies, and in vitro Characterization of Cinnamic Acid Derivatives as SmHDAC8 Inhibitors for the Treatment of Schistosomiasis.
ChemMedChem, 13, 2018
|
|
1ZWD
 
 | |
1ZWA
 
 | |
1ZWE
 
 | |
1ZWC
 
 | |
