6DM3
 
 | | Crystal structure of the SH2 domain from RavO (Lpg1129) from Legionella pneumophila, apoprotein | | Descriptor: | RavO | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Kaneko, T, Li, S, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2018-06-04 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the SH2 domain from RavO (Lpg1129) from Legionella pneumophila, apoprotein
To Be Published
|
|
6WJ8
 
 | | Crystal structure of gamma-aminobutyrate aminotransferase PuuE from Klebsiella pneumoniae in complex with PLP | | Descriptor: | 4-aminobutyrate aminotransferase PuuE | | Authors: | Stogios, P.J, Evdokimova, E, McChesney, C, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-13 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of gamma-aminobutyrate aminotransferase PuuE from Klebsiella pneumoniae in complex with PLP
To Be Published
|
|
5KVK
 
 | | Crystal structure of the Competence-Damaged Protein (CinA) Superfamily Protein KP700603 from Klebsiella pneumoniae 700603 | | Descriptor: | Protein KP700603 | | Authors: | Stogios, P.J, Wawrzak, Z, Evdokimova, E, Di Leo, R, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-14 | | Release date: | 2016-08-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | To be published
To Be Published
|
|
8V4U
 
 | |
6MN0
 
 | | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, H168A mutant in complex with acetyl-CoA | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ACETYL COENZYME *A, Aminoglycoside N(3)-acetyltransferase, ... | | Authors: | Stogios, P.J, Skarina, T, Zu, X, Yim, V, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
6MMZ
 
 | | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, H29A mutant apoenzyme | | Descriptor: | Aminoglycoside N(3)-acetyltransferase, CHLORIDE ION, SULFATE ION | | Authors: | Stogios, P.J, Skarina, T, Xu, Z, Yim, V, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
7UUJ
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with gentamicin | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 1,2-ETHANEDIOL, Aminocyclitol acetyltransferase ApmA, ... | | Authors: | Stogios, P.J, Evdokimova, E, Osipiuk, J, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-04-28 | | Release date: | 2023-04-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with gentamicin
To Be Published
|
|
6VU7
 
 | | Crystal structure of YbjN, a putative transcription regulator from E. coli | | Descriptor: | CHLORIDE ION, YbjN protein | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-02-14 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of YbjN, a putative transcription regulator from E. coli
To Be Published
|
|
6E8M
 
 | |
6E8H
 
 | | Legionella Longbeachae LeSH (Llo2327) | | Descriptor: | CHLORIDE ION, LeSH (Llo2327) | | Authors: | Kaneko, T, Li, S.S.C. | | Deposit date: | 2018-07-29 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Identification and characterization of a large family of superbinding bacterial SH2 domains.
Nat Commun, 9, 2018
|
|
6E8K
 
 | |
6E8I
 
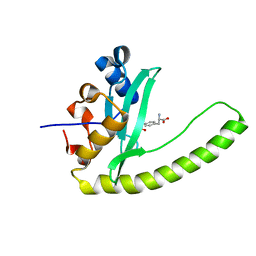 | |
5HT0
 
 | | Crystal structure of an Antibiotic_NAT family aminoglycoside acetyltransferase HMB0038 from an uncultured soil metagenomic sample in complex with coenzyme A | | Descriptor: | Aminoglycoside acetyltransferase HMB0005, COENZYME A, SULFATE ION | | Authors: | Xu, Z, Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-26 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.752 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
7KES
 
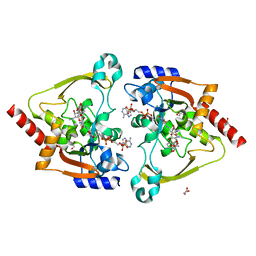 | | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, mutant H168A in complex with apramycin and CoA | | Descriptor: | APRAMYCIN, Aminoglycoside N(3)-acetyltransferase, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Skarina, T, Michalska, K, Xu, Z, Yim, V, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-10-12 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
3S8Y
 
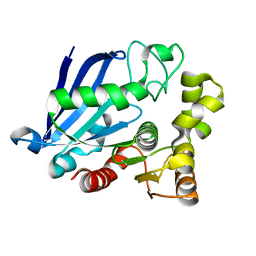 | | Bromide soaked structure of an esterase from the oil-degrading bacterium Oleispira antarctica | | Descriptor: | BROMIDE ION, Esterase APC40077 | | Authors: | Petit, P, Dong, A, Kagan, O, Savchenko, A, Yakunin, A.F. | | Deposit date: | 2011-05-31 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and activity of the cold-active and anion-activated carboxyl esterase OLEI01171 from the oil-degrading marine bacterium Oleispira antarctica.
Biochem.J., 445, 2012
|
|
6VTV
 
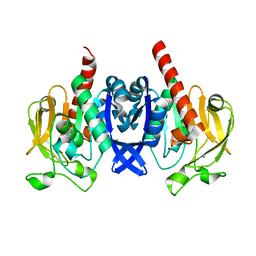 | | Crystal structure of PuuD gamma-glutamyl-gamma-aminobutyrate hydrolase from E. coli | | Descriptor: | Gamma-glutamyl-gamma-aminobutyrate hydrolase PuuD, MANGANESE (II) ION | | Authors: | Stogios, P.J, EVDOKIMOVA, E, DI LEO, R, SAVCHENKO, A, JOACHIMIAK, A, SATCHELL, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-02-13 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | PuuD gamma-glutamyl-gamma-aminobutyrate hydrolase
To Be Published
|
|
5SUJ
 
 | | Crystal structure of uncharacterized protein LPG2148 from Legionella pneumophila | | Descriptor: | Uncharacterized protein | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-08-03 | | Release date: | 2016-08-17 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.356 Å) | | Cite: | Discovery of Ubiquitin Deamidases in the Pathogenic Arsenal of Legionella pneumophila.
Cell Rep, 23, 2018
|
|
7MX5
 
 | | Crystal structure of TolB from Acinetobacter baumannii | | Descriptor: | Tol-Pal system protein TolB | | Authors: | Stogios, P.J, Evdokimova, E, Endres, M, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-18 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystal structure of TolB from Acinetobacter baumannii
To Be Published
|
|
6XI5
 
 | |
6XI4
 
 | |
7RCA
 
 | | Crystal structure of Aro2p chorismate synthase from Candida lusitaniae | | Descriptor: | CHLORIDE ION, Chorismate synthase, SULFATE ION | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-07 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of Aro2p chorismate synthase from Candida lusitaniae
To Be Published
|
|
7RGN
 
 | | Crystal structure of putative fructose-1,6-bisphosphate aldolase from Candida auris | | Descriptor: | Fructose-bisphosphate aldolase, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Stogios, P.J, Evdokimova, E, Tan, K, Di Leo, R, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-15 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of putative fructose-1,6-bisphosphate aldolase from Candida auris
To Be Published
|
|
7REU
 
 | | Crystal structure of Aro4p, 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) synthase from Candida auris, L-Tyr complex | | Descriptor: | 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) synthase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Evdokimova, E, Tan, K, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-13 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of Aro4p, 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) synthase from Candida auris, L-Tyr complex
To Be Published
|
|
7RGE
 
 | | Crystal structure of phosphoadenylyl-sulfate (PAPS) reductase from Candida auris, phosphate complex | | Descriptor: | 3'-phosphoadenylylsulfate reductase, GLYCEROL, PHOSPHATE ION | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-15 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of phosphoadenylyl-sulfate (PAPS) reductase from Candida auris, phosphate complex
To Be Published
|
|
7RJ1
 
 | | Crystal structure of Aro7p chorismate mutase from Candida albicans, complex with L-Trp | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Chorismate mutase, ... | | Authors: | Stogios, P.J, Evdokimova, E, Tan, K, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-20 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of Aro7p chorismate mutase from Candida albicans, complex with L-Trp
To Be Published
|
|
