8IFS
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 7 | | Descriptor: | (8~{S})-7-[(2~{S})-2-(~{tert}-butylcarbamoylamino)-3,3-dimethyl-butanoyl]-~{N}-[(1~{S})-1-cyano-2-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]ethyl]-1,4-dithia-7-azaspiro[4.4]nonane-8-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IFT
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 10 | | Descriptor: | (8S)-N-[(1S)-1-cyano-2-[(3S)-2-oxidanylidenepyrrolidin-3-yl]ethyl]-7-[(2S)-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-1,4-dithia-7-azaspiro[4.4]nonane-8-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IFR
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 3 | | Descriptor: | (1R,2S,5S)-3-[(2S)-2-(tert-butylcarbamoylamino)-3,3-dimethyl-butanoyl]-6,6-dimethyl-N-[(2S)-5-oxidanylidene-1-[(3S)-2-oxidanylidenepyrrolidin-3-yl]hex-3-en-2-yl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IGX
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 9 (simnotrelvir, SIM0417, SSD8432) | | Descriptor: | (8~{S})-~{N}-[(1~{S})-1-cyano-2-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]ethyl]-7-[(2~{S})-3,3-dimethyl-2-[2,2,2-tris(fluoranyl)ethanoylamino]butanoyl]-1,4-dithia-7-azaspiro[4.4]nonane-8-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IGY
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
7DC3
 
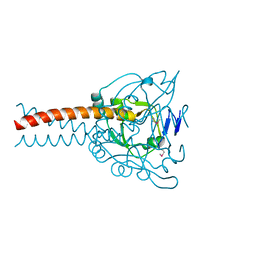 | | Crystal structure of the MyRF ICA domain | | Descriptor: | Isoform 2 of Myelin regulatory factor | | Authors: | Shi, N, Wu, P, Zhen, X.K, Li, B.W. | | Deposit date: | 2020-10-23 | | Release date: | 2021-08-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the MyRF ICA domain with its upstream beta-helical stalk reveals the molecular mechanisms underlying its trimerization and self-cleavage.
Int J Biol Sci, 17, 2021
|
|
3JCO
 
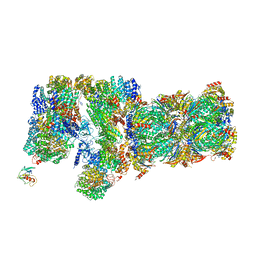 | | Structure of yeast 26S proteasome in M1 state derived from Titan dataset | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Luan, B, Huang, X.L, Wu, J.P, Shi, Y.G, Wang, F. | | Deposit date: | 2016-01-06 | | Release date: | 2016-06-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure of an endogenous yeast 26S proteasome reveals two major conformational states.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JFB
 
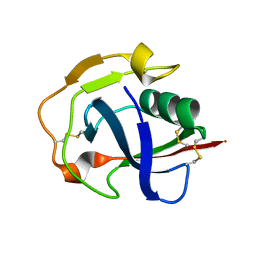 | | Crystal structure of the scavenger receptor cysteine-rich domain 5 (SRCR5) from porcine CD163 | | Descriptor: | Scavenger receptor cysteine-rich type 1 protein M130 | | Authors: | Ma, H, Jiang, L, Qiao, S, Zhang, G, Li, R. | | Deposit date: | 2016-04-19 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of the Fifth Scavenger Receptor Cysteine-Rich Domain of Porcine CD163 Reveals an Important Residue Involved in Porcine Reproductive and Respiratory Syndrome Virus Infection
J. Virol., 91, 2017
|
|
4IAR
 
 | | Crystal structure of the chimeric protein of 5-HT1B-BRIL in complex with ergotamine (PSI Community Target) | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chimera protein of human 5-hydroxytryptamine receptor 1B and E. Coli soluble cytochrome b562, Ergotamine | | Authors: | Wang, C, Jiang, Y, Ma, J, Wu, H, Wacker, D, Katritch, V, Han, G.W, Liu, W, Huang, X, Vardy, E, McCorvy, J.D, Gao, X, Zhou, E.X, Melcher, K, Zhang, C, Bai, F, Yang, H, Yang, L, Jiang, H, Roth, B.L, Cherezov, V, Stevens, R.C, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for molecular recognition at serotonin receptors.
Science, 340, 2013
|
|
7TJE
 
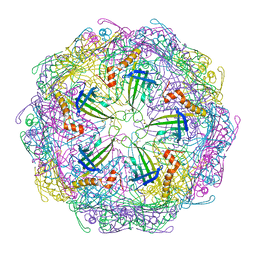 | | Bacteriophage Q beta capsid protein A38K | | Descriptor: | Minor capsid protein A1 | | Authors: | Jin, X. | | Deposit date: | 2022-01-16 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Alternative Assembly of Q beta Virus-like Particles
To Be Published
|
|
7TJG
 
 | |
7TJM
 
 | |
7TJD
 
 | |
4IAQ
 
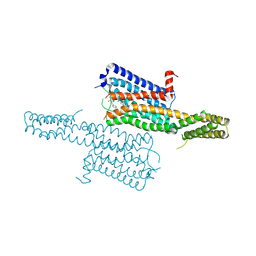 | | Crystal structure of the chimeric protein of 5-HT1B-BRIL in complex with dihydroergotamine (PSI Community Target) | | Descriptor: | Chimera protein of human 5-hydroxytryptamine receptor 1B and E. Coli soluble cytochrome b562, Dihydroergotamine | | Authors: | Wang, C, Jiang, Y, Ma, J, Wu, H, Wacker, D, Katritch, V, Han, G.W, Liu, W, Huang, X, Vardy, E, McCorvy, J.D, Gao, X, Zhou, E.X, Melcher, K, Zhang, C, Bai, F, Yang, H, Yang, L, Jiang, H, Roth, B.L, Cherezov, V, Stevens, R.C, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for molecular recognition at serotonin receptors.
Science, 340, 2013
|
|
6VIZ
 
 | | BRD4_Bromodomain1 complex with pyrrolopyridone compound 27 | | Descriptor: | 4-[2-(2,6-dimethylphenoxy)-5-(ethylsulfonyl)phenyl]-N-ethyl-6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide, Bromodomain-containing protein 4 | | Authors: | Longenecker, K.L, Park, C.H, Qiu, W. | | Deposit date: | 2020-01-14 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.391 Å) | | Cite: | Discovery ofN-Ethyl-4-[2-(4-fluoro-2,6-dimethyl-phenoxy)-5-(1-hydroxy-1-methyl-ethyl)phenyl]-6-methyl-7-oxo-1H-pyrrolo[2,3-c]pyridine-2-carboxamide (ABBV-744), a BET Bromodomain Inhibitor with Selectivity for the Second Bromodomain.
J.Med.Chem., 63, 2020
|
|
6VIW
 
 | | BRD4_Bromodomain1 complex with pyrrolopyridone compound 18 | | Descriptor: | 4-[2-(2,4-difluorophenoxy)-5-(methylsulfonyl)phenyl]-N-ethyl-6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide, Bromodomain-containing protein 4 | | Authors: | Longenecker, K.L, Park, C.H, Qiu, W. | | Deposit date: | 2020-01-14 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.429 Å) | | Cite: | Discovery ofN-Ethyl-4-[2-(4-fluoro-2,6-dimethyl-phenoxy)-5-(1-hydroxy-1-methyl-ethyl)phenyl]-6-methyl-7-oxo-1H-pyrrolo[2,3-c]pyridine-2-carboxamide (ABBV-744), a BET Bromodomain Inhibitor with Selectivity for the Second Bromodomain.
J.Med.Chem., 63, 2020
|
|
6VIX
 
 | | BRD4_Bromodomain2 complex with pyrrolopyridone compound 18 | | Descriptor: | 4-[2-(2,4-difluorophenoxy)-5-(methylsulfonyl)phenyl]-N-ethyl-6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide, Bromodomain-containing protein 4 | | Authors: | Longenecker, K.L, Park, C.H, Qiu, W. | | Deposit date: | 2020-01-14 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.116 Å) | | Cite: | Discovery ofN-Ethyl-4-[2-(4-fluoro-2,6-dimethyl-phenoxy)-5-(1-hydroxy-1-methyl-ethyl)phenyl]-6-methyl-7-oxo-1H-pyrrolo[2,3-c]pyridine-2-carboxamide (ABBV-744), a BET Bromodomain Inhibitor with Selectivity for the Second Bromodomain.
J.Med.Chem., 63, 2020
|
|
6ZGC
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound Saracatinib (AZD0530) | | Descriptor: | Activin receptor type I, N-(5-CHLORO-1,3-BENZODIOXOL-4-YL)-7-[2-(4-METHYLPIPERAZIN-1-YL)ETHOXY]-5-(TETRAHYDRO-2H-PYRAN-4-YLOXY)QUINAZOLIN-4-AMINE, PHOSPHATE ION, ... | | Authors: | Williams, E.P, Galan Bartual, S, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-06-18 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Saracatinib is an efficacious clinical candidate for fibrodysplasia ossificans progressiva.
JCI Insight, 6, 2021
|
|
6VIY
 
 | | BRD2_Bromodomain2 complex with pyrrolopyridone compound 27 | | Descriptor: | 4-[2-(2,6-dimethylphenoxy)-5-(ethylsulfonyl)phenyl]-N-ethyl-6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide, Bromodomain-containing protein 2 | | Authors: | Longenecker, K.L, Park, C.H, Qiu, W. | | Deposit date: | 2020-01-14 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Discovery ofN-Ethyl-4-[2-(4-fluoro-2,6-dimethyl-phenoxy)-5-(1-hydroxy-1-methyl-ethyl)phenyl]-6-methyl-7-oxo-1H-pyrrolo[2,3-c]pyridine-2-carboxamide (ABBV-744), a BET Bromodomain Inhibitor with Selectivity for the Second Bromodomain.
J.Med.Chem., 63, 2020
|
|
4REY
 
 | | Crystal Structure of the GRASP65-GM130 C-terminal peptide complex | | Descriptor: | Golgi reassembly-stacking protein 1, Golgin subfamily A member 2, SULFATE ION | | Authors: | Shi, N, Hu, F, Li, B. | | Deposit date: | 2014-09-24 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural Basis for the Interaction between the Golgi Reassembly-stacking Protein GRASP65 and the Golgi Matrix Protein GM130.
J.Biol.Chem., 290, 2015
|
|
7CR5
 
 | |
6M1H
 
 | | CryoEM structure of human PAC1 receptor in complex with maxadilan | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Song, X, Wang, J, Zhang, D, Wang, H.W, Ma, Y. | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-11 | | Last modified: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of PAC1 receptor reveal ligand binding mechanism.
Cell Res., 30, 2020
|
|
6M1I
 
 | | CryoEM structure of human PAC1 receptor in complex with PACAP38 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Song, X, Wang, J, Zhang, D, Wang, H.W, Ma, Y. | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-11 | | Last modified: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of PAC1 receptor reveal ligand binding mechanism.
Cell Res., 30, 2020
|
|
5J8V
 
 | |
5UEX
 
 | | BRD4_BD2_A-1497627 | | Descriptor: | 17-{[ethyl(dihydroxy)-lambda~4~-sulfanyl]amino}-11,13-difluoro-2-methyl-6,7,8,9-tetrahydrodibenzo[4,5:7,8][1,6]dioxacyclododecino[3,2-c]pyridin-3(2H)-one, Bromodomain-containing protein 4 | | Authors: | Park, C.H. | | Deposit date: | 2017-01-03 | | Release date: | 2017-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Fragment-Based, Structure-Enabled Discovery of Novel Pyridones and Pyridone Macrocycles as Potent Bromodomain and Extra-Terminal Domain (BET) Family Bromodomain Inhibitors.
J. Med. Chem., 60, 2017
|
|
