6Y2P
 
 | |
6YJM
 
 | | Crystal Structure of the Catalytic Domain of ADAMTS-5 in Complex with the Inhibitor GLPG1972 | | Descriptor: | (5~{S})-5-[3-[(3~{S})-4-[3,5-bis(fluoranyl)phenyl]-3-methyl-piperazin-1-yl]-3-oxidanylidene-propyl]-5-cyclopropyl-imidazolidine-2,4-dione, A disintegrin and metalloproteinase with thrombospondin motifs 5, CALCIUM ION, ... | | Authors: | Goepfert, A, Leonard, P, Triballeau, N, Fleury, D, Mollat, P, Lamers, M. | | Deposit date: | 2020-04-03 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of GLPG1972/S201086, a Potent, Selective, and Orally Bioavailable ADAMTS-5 Inhibitor for the Treatment of Osteoarthritis.
J.Med.Chem., 64, 2021
|
|
6T9I
 
 | | cryo-EM structure of transcription coactivator SAGA | | Descriptor: | Protein SPT3, SAGA-associated factor 73, Transcription factor SPT20, ... | | Authors: | Wang, H, Cheung, A, Cramer, P. | | Deposit date: | 2019-10-28 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the transcription coactivator SAGA.
Nature, 577, 2020
|
|
8A5R
 
 | | Crystal structure of light-activated DNA-binding protein EL222 from Erythrobacter litoralis crystallized and measured in dark. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Koval, T, Chaudhari, A, Fuertes, G, Andersson, I, Dohnalek, J. | | Deposit date: | 2022-06-15 | | Release date: | 2023-07-05 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Light-dependent flavin redox and adduct states control the conformation and DNA-binding activity of the transcription factor EL222.
Nucleic Acids Res., 53, 2025
|
|
6T3V
 
 | | Psychrophilic aromatic amino acids aminotransferase from Psychrobacter sp. B6 cocrystalized with substrate analog - malic acid | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Rutkiewicz, M, Bujacz, A, Rum, J, Bujacz, G. | | Deposit date: | 2019-10-11 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural Evidence of Active Site Adaptability towards Different Sized Substrates of Aromatic Amino Acid Aminotransferase from Psychrobacter Sp. B6.
Materials, 14, 2021
|
|
8A4D
 
 | |
6TGU
 
 | | Crystal structure of human protein kinase CK2alpha'(CSNK2A2 gene product) in complex with the 2-aminothiazole-type inhibitor Cl-OH-3 | | Descriptor: | 1,2-ETHANEDIOL, 4-[[4-(4-chlorophenyl)-1,3-thiazol-2-yl]amino]-2-oxidanyl-benzoic acid, Casein kinase II subunit alpha' | | Authors: | Niefind, K, Lindenblatt, D, Jose, J, Applegate, V.M, Nickelsen, A. | | Deposit date: | 2019-11-18 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (0.833 Å) | | Cite: | Structural and Mechanistic Basis of the Inhibitory Potency of Selected 2-Aminothiazole Compounds on Protein Kinase CK2.
J.Med.Chem., 63, 2020
|
|
8A45
 
 | |
8A5K
 
 | |
8A5S
 
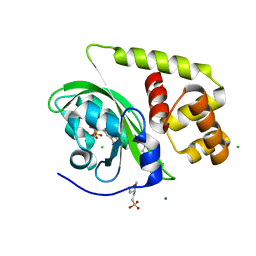 | | Crystal structure of light-activated DNA-binding protein EL222 from Erythrobacter litoralis crystallized in dark, measured illuminated. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Koval, T, Chaudhari, A, Fuertes, G, Andersson, I, Dohnalek, J. | | Deposit date: | 2022-06-15 | | Release date: | 2023-07-05 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Light-dependent flavin redox and adduct states control the conformation and DNA-binding activity of the transcription factor EL222.
Nucleic Acids Res., 53, 2025
|
|
6YQ5
 
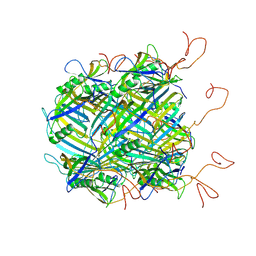 | | Hybrid structure of the SPP1 tail tube by solid-state NMR and cryo EM - NMR Ensemble | | Descriptor: | Tail tube protein gp17.1* | | Authors: | Zinke, M, Sachowsky, K.A.A, Zinn-Justin, S, Ravelli, R, Schroder, G.F, Habeck, M, Lange, A. | | Deposit date: | 2020-04-16 | | Release date: | 2020-10-14 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4 Å), SOLID-STATE NMR | | Cite: | Architecture of the flexible tail tube of bacteriophage SPP1.
Nat Commun, 11, 2020
|
|
6YE5
 
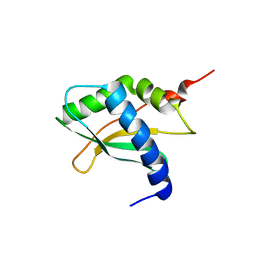 | | Structure of ribosomal binding factor A RbfA of Staphylococcus aureus bacterium by NMR | | Descriptor: | Ribosome-binding factor A | | Authors: | Blokhin, D.S, Usachev, K.S, Bikmullin, A.G, Nurullina, L, Garaeva, N, Validov, S, Klochkov, V, Aganov, A, Khusainov, I, Yusupov, M. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure of ribosomal binding factor A RbfA of Staphylococcus aureus bacterium by NMR
To Be Published
|
|
6T72
 
 | | Structure of the RsaA N-terminal domain bound to LPS | | Descriptor: | 4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose, CALCIUM ION, S-layer protein | | Authors: | von Kuegelgen, A, Bharat, T.A.M. | | Deposit date: | 2019-10-21 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | In Situ Structure of an Intact Lipopolysaccharide-Bound Bacterial Surface Layer.
Cell, 180, 2020
|
|
6Z1M
 
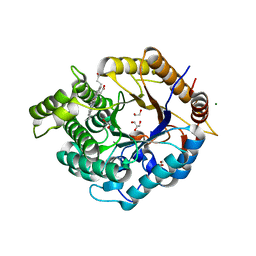 | | Structure of an Ancestral glycosidase (family 1) bound to heme | | Descriptor: | 1,2-ETHANEDIOL, Ancestral reconstructed glycosidase, GLYCEROL, ... | | Authors: | Gavira, J.A, Risso, V.A, Sanchez-Ruiz, J.M, Gamiz-Arco, G, Gutierrez-Rus, L, Ibarra-Molero, B, Oshino, Y, Petrovic, D, Romero-Rivera, A, Seelig, B, Kamerlin, S.C.L, Gaucher, E.A. | | Deposit date: | 2020-05-14 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Heme-binding enables allosteric modulation in an ancient TIM-barrel glycosidase.
Nat Commun, 12, 2021
|
|
6Z7P
 
 | | Composite model of the Caulobacter crescentus S-layer bound to the O-antigen of lipopolysaccharide | | Descriptor: | 4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose, CALCIUM ION, S-layer protein | | Authors: | Bharat, T.A.M, von Kugelgen, A. | | Deposit date: | 2020-06-01 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | In Situ Structure of an Intact Lipopolysaccharide-Bound Bacterial Surface Layer.
Cell, 180, 2020
|
|
8ILU
 
 | | Crystal structure of mouse Galectin-3 in complex with small molecule inhibitor | | Descriptor: | (2R,3R,4R,5R,6S)-2-(hydroxymethyl)-6-[2-(2-methyl-1,3-benzothiazol-6-yl)-1,2,4-triazol-3-yl]-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, Galectin-3, SODIUM ION, ... | | Authors: | Kumar, A, Jinal, S, Raman, S, Ghosh, K. | | Deposit date: | 2023-03-04 | | Release date: | 2024-03-06 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of benzothiazole derived monosaccharides as potent, selective, and orally bioavailable inhibitors of human and mouse galectin-3; a rare example of using a S···O binding interaction for drug design.
Bioorg.Med.Chem., 101, 2024
|
|
6TIT
 
 | | VSV G_440 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Albertini, A.A, Belot, L, Abouhamdan, A, Gaudin, Y. | | Deposit date: | 2019-11-22 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Identification of a pH-Sensitive Switch in VSV-G and a Crystal Structure of the G Pre-fusion State Highlight the VSV-G Structural Transition Pathway.
Cell Rep, 32, 2020
|
|
6TDA
 
 | | Structure of SWI/SNF chromatin remodeler RSC bound to a nucleosome | | Descriptor: | Actin-like protein ARP9, Actin-related protein 7, Chromatin structure-remodeling complex protein RSC58, ... | | Authors: | Wagner, F.R, Dienemann, C, Wang, H, Stuetzer, A, Tegunov, D, Urlaub, H, Cramer, P. | | Deposit date: | 2019-11-08 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Structure of SWI/SNF chromatin remodeller RSC bound to a nucleosome.
Nature, 579, 2020
|
|
8B1N
 
 | | Crystal structure of TrmD-Tm1570 from Calditerrivibrio nitroreducens in complex with S-adenosyl-L-methionine | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kluza, A, Lewandowska, I, Augustyniak, R, Sulkowska, J. | | Deposit date: | 2022-09-10 | | Release date: | 2022-09-28 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Are there double knots in proteins? Prediction and in vitro verification based on TrmD-Tm1570 fusion from C. nitroreducens.
Front Mol Biosci, 10, 2023
|
|
7CI7
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | (2R,3R)-2-azanyl-1-[4-[[4-[2-[4-(hydroxymethyl)phenyl]ethynyl]phenyl]methyl]piperidin-1-yl]-4-methylsulfonyl-3-oxidanyl-butan-1-one, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Mima, M, Baker, L.M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
7CI6
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | (1S)-1-[1-[3-(4-chlorophenyl)propyl]imidazol-2-yl]ethanol, CHLORIDE ION, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Mima, M, Baker, L.M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
7CI8
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | (1S)-1-[1-[(5-phenyl-1,2-oxazol-3-yl)methyl]imidazol-2-yl]ethanol, MAGNESIUM ION, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Mima, M, Baker, L.M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
7CIE
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | (2R)-2-azanyl-3-oxidanyl-N-[3-(trifluoromethyloxy)phenyl]propanamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Baker, L.M, Mima, M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
8B7J
 
 | | Human HSP90 alpha ATP Binding Domain, ATP-lid closed conformation, R46A | | Descriptor: | HSP90AA1 protein | | Authors: | Rioual, E, Henot, F, Favier, A, Macek, P, Crublet, E, Josso, P, Brustcher, B, Frech, M, Gans, P, Loison, C, Boisbouvier, J. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Visualizing the transiently populated closed-state of human HSP90 ATP binding domain.
Nat Commun, 13, 2022
|
|
8B7I
 
 | | Human HSP90 alpha ATP Binding Domain, ATP-lid open conformation, R60A | | Descriptor: | HSP90AA1 protein | | Authors: | Rioual, E, Henot, F, Favier, A, Macek, P, Crublet, E, Josso, P, Brutscher, B, Frech, M, Gans, P, Loison, C, Boisbouvier, J. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Visualizing the transiently populated closed-state of human HSP90 ATP binding domain.
Nat Commun, 13, 2022
|
|
