7FBK
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain N501Y mutant in complex with neutralizing nanobody 20G6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, New antigen receptor variable domain, Spike protein S1 | | Authors: | Zhu, J, Xu, T, Feng, B, Liu, J. | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Class of Shark-Derived Single-Domain Antibodies can Broadly Neutralize SARS-Related Coronaviruses and the Structural Basis of Neutralization and Omicron Escape.
Small Methods, 6, 2022
|
|
6L7P
 
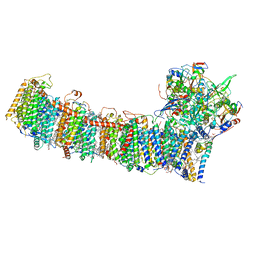 | | cryo-EM structure of cyanobacteria NDH-1LdelV complex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, BETA-CAROTENE, ... | | Authors: | Zhang, C, Shuai, J, Wu, J, Lei, M. | | Deposit date: | 2019-11-02 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into NDH-1 mediated cyclic electron transfer.
Nat Commun, 11, 2020
|
|
1R9H
 
 | |
7F46
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab (state1, local refinement of the RBD, NTD and 35B5 Fab) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-06-17 | | Release date: | 2022-03-23 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.79 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7FJO
 
 | | Cryo-EM structure of South African (B.1.351) SARS-CoV-2 spike glycoprotein in complex with three T6 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, T6 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Zhang, S, Liang, Q. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | RBD trimer mRNA vaccine elicits broad and protective immune responses against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
7FJS
 
 | | Crystal structure of T6 Fab bound to theSARS-CoV-2 RBD of B.1.351 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, T6 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Zhang, S, Liang, Q. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | RBD trimer mRNA vaccine elicits broad and protective immune responses against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
7FJN
 
 | | Cryo-EM structure of South African (B.1.351) SARS-CoV-2 spike glycoprotein in complex with two T6 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Envelope glycoprotein, T6 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Zhang, S, Liang, Q. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | RBD trimer mRNA vaccine elicits broad and protective immune responses against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
5VXA
 
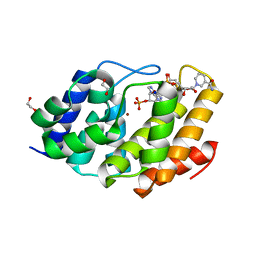 | | Structure of the human Mesh1-NADPH complex | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Rose, J, Zhou, P. | | Deposit date: | 2017-05-23 | | Release date: | 2018-05-23 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | MESH1 is a cytosolic NADPH phosphatase that regulates ferroptosis.
Nat Metab, 2, 2020
|
|
6J2W
 
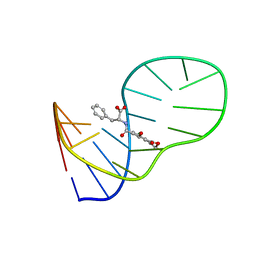 | | The structure of OBA3-OTA complex | | Descriptor: | DNA (5'-D(*CP*GP*GP*GP*GP*CP*GP*AP*AP*GP*CP*GP*GP*GP*TP*CP*CP*CP*G)-3'), N-[(3R)-5-chloro-8-hydroxy-3-methyl-1-oxo-3,4-dihydro-1H-2-benzopyran-7-carbonyl]-D-phenylalanine | | Authors: | Xu, G.H, Li, C.G. | | Deposit date: | 2019-01-03 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure-guided post-SELEX optimization of an ochratoxin A aptamer.
Nucleic Acids Res., 47, 2019
|
|
6SJ7
 
 | |
7EPX
 
 | | S protein of SARS-CoV-2 in complex with GW01 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Shen, Y.P, Zhang, Y.Y, Yan, R.H, Li, Y.N, Zhou, Q. | | Deposit date: | 2021-04-28 | | Release date: | 2022-05-18 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Novel sarbecovirus bispecific neutralizing antibodies with exceptional breadth and potency against currently circulating SARS-CoV-2 variants and sarbecoviruses.
Cell Discov, 8, 2022
|
|
5E00
 
 | | Structure of HLA-A2 P130 | | Descriptor: | Beta-2-microglobulin, GLY-VAL-TRP-ILE-ARG-THR-PRO-PRO-ALA, HLA class I histocompatibility antigen, ... | | Authors: | Zhang, Y, Wu, Y, Qi, J, Liu, J, Gao, G.F, Meng, S. | | Deposit date: | 2015-09-26 | | Release date: | 2017-01-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | CD8+T-Cell Response-Associated Evolution of Hepatitis B Virus Core Protein and Disease Progress.
J. Virol., 92, 2018
|
|
5B25
 
 | | Crystal structure of human PDE1B with inhibitor 3 | | Descriptor: | (11R,15S)-4-{[4-(6-fluoropyridin-2-yl)phenyl]methyl}-8-methyl-5-(phenylamino)-1,3,4,8,10-pentaazatetracyclo[7.6.0.02,6.011,15]pentadeca-2,5,9-trien-7-one, Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B, GLYCEROL, ... | | Authors: | Ida, K, Lane, W, Snell, G, Sogabe, S. | | Deposit date: | 2016-01-07 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Potent and Selective Inhibitors of Phosphodiesterase 1 for the Treatment of Cognitive Impairment Associated with Neurodegenerative and Neuropsychiatric Diseases
J.Med.Chem., 59, 2016
|
|
5VYY
 
 | |
9KR5
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with compound 3 | | Descriptor: | (6~{E})-6-(6-chloranyl-2-methyl-indazol-5-yl)imino-3-[5-[(3~{S})-oxolan-3-yl]oxypyridin-3-yl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazinane-2,4-dione, 3C-like proteinase nsp5 | | Authors: | Zhong, Y, Zhao, L, Zhang, W, Peng, W. | | Deposit date: | 2024-11-27 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Expanding the utilization of binding pockets proves to be effective for noncovalent small molecule inhibitors against SARS-CoV-2 M pro.
Eur.J.Med.Chem., 289, 2025
|
|
9KSK
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with compound 10 | | Descriptor: | 3C-like proteinase, 4-[4-chloranyl-2-[[(6E)-6-(6-chloranyl-2-methyl-indazol-5-yl)imino-3-(5-methylpyridin-3-yl)-2,4-bis(oxidanylidene)-1,3,5-triazinan-1-yl]methyl]-5-fluoranyl-phenoxy]-2-fluoranyl-benzenecarbonitrile | | Authors: | Zhong, Y, Zhao, L, Zhang, W, Peng, W. | | Deposit date: | 2024-11-29 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Expanding the utilization of binding pockets proves to be effective for noncovalent small molecule inhibitors against SARS-CoV-2 M pro.
Eur.J.Med.Chem., 289, 2025
|
|
9KSI
 
 | | Crystal Structure of SARS-CoV-2 main protease in complex with compound 5 | | Descriptor: | (6E)-1-[[5-chloranyl-4-fluoranyl-2-(4-fluoranylphenoxy)phenyl]methyl]-6-(6-chloranyl-2-methyl-indazol-5-yl)imino-3-(5-methoxypyridin-3-yl)-1,3,5-triazinane-2,4-dione, 3C-like proteinase nsp5 | | Authors: | Zhong, Y, Zhao, L, Zhang, W, Peng, W. | | Deposit date: | 2024-11-29 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Expanding the utilization of binding pockets proves to be effective for noncovalent small molecule inhibitors against SARS-CoV-2 M pro.
Eur.J.Med.Chem., 289, 2025
|
|
9KSH
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with compound 1 | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-pyridin-3-yl-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Zhong, Y, Zhao, L, Zhang, W, Peng, W. | | Deposit date: | 2024-11-29 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Expanding the utilization of binding pockets proves to be effective for noncovalent small molecule inhibitors against SARS-CoV-2 M pro.
Eur.J.Med.Chem., 289, 2025
|
|
9KSJ
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with compound 8 | | Descriptor: | 3-[[(6E)-6-(6-chloranyl-2-methyl-indazol-5-yl)imino-3-[5-(2-methoxyethoxy)pyridin-3-yl]-2,4-bis(oxidanylidene)-1,3,5-triazinan-1-yl]methyl]-4-methyl-benzenecarbonitrile, 3C-like proteinase nsp5 | | Authors: | Zhong, Y, Zhao, L, Zhang, W, Peng, W. | | Deposit date: | 2024-11-29 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Expanding the utilization of binding pockets proves to be effective for noncovalent small molecule inhibitors against SARS-CoV-2 M pro.
Eur.J.Med.Chem., 289, 2025
|
|
5WMT
 
 | |
5GSX
 
 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide 142-2 | | Descriptor: | 10-mer peptide from Spike protein, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-17 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
5BKH
 
 | |
4MK7
 
 | |
4MK8
 
 | | Hepatitis C Virus polymerase NS5B genotype 1b (BK) in complex with inhibitor 4 (N-(4-{2-[3-tert-butyl-2-methoxy-5-(2-oxo-1,2-dihydropyridin-3-yl)phenyl]ethyl}phenyl)methanesulfonamide) | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, N-(4-{2-[3-tert-butyl-2-methoxy-5-(2-oxo-1,2-dihydropyridin-3-yl)phenyl]ethyl}phenyl)methanesulfonamide, ... | | Authors: | Harris, S.F, Wong, A. | | Deposit date: | 2013-09-04 | | Release date: | 2013-10-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of a Novel Series of Potent Non-Nucleoside Inhibitors of Hepatitis C Virus NS5B.
J.Med.Chem., 56, 2013
|
|
4MKB
 
 | |
