2O6L
 
 | |
6HTF
 
 | |
2LOU
 
 | | AR55 solubilised in DPC micelles | | Descriptor: | Apelin receptor | | Authors: | Langelaan, D.N, Rainey, J.K. | | Deposit date: | 2012-01-27 | | Release date: | 2013-01-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural features of the apelin receptor N-terminal tail and first transmembrane segment implicated in ligand binding and receptor trafficking.
Biochim.Biophys.Acta, 1828, 2013
|
|
4L00
 
 | |
4L01
 
 | |
3FPD
 
 | | G9a-like protein lysine methyltransferase inhibition by BIX-01294 | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5, N-(1-benzylpiperidin-4-yl)-6,7-dimethoxy-2-(4-methyl-1,4-diazepan-1-yl)quinazolin-4-amine, ... | | Authors: | Chang, Y, Zhang, X, Horton, J.R, Cheng, X. | | Deposit date: | 2009-01-05 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294.
Nat.Struct.Mol.Biol., 16, 2009
|
|
4RBU
 
 | |
4RFW
 
 | | Crystal structure of human retinoid X Receptor alpha-ligand binding domain complex with 9cUAB70 and the coactivator peptide GRIP-1 | | Descriptor: | (2E,4E,6Z,8E)-3,7-dimethyl-8-(6,7,8,9-tetrahydro-5H-benzo[7]annulen-5-ylidene)octa-2,4,6-trienoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Xia, G, Muccio, D.D, Smith, C.D. | | Deposit date: | 2014-09-28 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformationally Defined Rexinoids and Their Efficacy in the Prevention of Mammary Cancers.
J.Med.Chem., 58, 2015
|
|
4RME
 
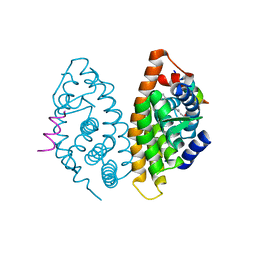 | | Crystal structure of human Retinoid X receptor alpha ligand binding domain complex with 9cUAB111 and coactivator peptide GRIP-1 | | Descriptor: | (2E,4E,6Z,8E)-3,7-dimethyl-8-[2-(3-methylbutyl)-3-(propan-2-yl)cyclohex-2-en-1-ylidene]octa-2,4,6-trienoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Xia, G, Muccio, D.D. | | Deposit date: | 2014-10-21 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformationally Defined Rexinoids and Their Efficacy in the Prevention of Mammary Cancers.
J.Med.Chem., 58, 2015
|
|
4RBT
 
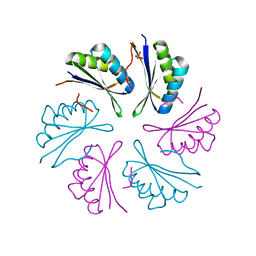 | |
4RMD
 
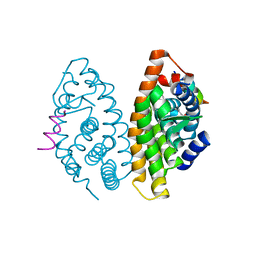 | | Crystal structure of human Retinoid X receptor alpha ligand binding domain complex with 9cUAB110 and coactivator peptide GRIP-1 | | Descriptor: | (2E,4E,6Z,8E)-8-[3-cyclopropyl-2-(3-methylbutyl)cyclohex-2-en-1-ylidene]-3,7-dimethylocta-2,4,6-trienoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Xia, G, Muccio, D.D. | | Deposit date: | 2014-10-21 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformationally Defined Rexinoids and Their Efficacy in the Prevention of Mammary Cancers.
J.Med.Chem., 58, 2015
|
|
3HUP
 
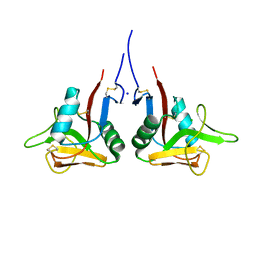 | | High-resolution structure of the extracellular domain of human CD69 | | Descriptor: | CHLORIDE ION, Early activation antigen CD69, SODIUM ION | | Authors: | Kolenko, P, Dohnalek, J, Skalova, T, Hasek, J, Duskova, J, Vanek, O, Bezouska, K. | | Deposit date: | 2009-06-15 | | Release date: | 2009-12-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.371 Å) | | Cite: | The high-resolution structure of the extracellular domain of human CD69 using a novel polymer
Acta Crystallogr.,Sect.F, 65, 2009
|
|
4RMC
 
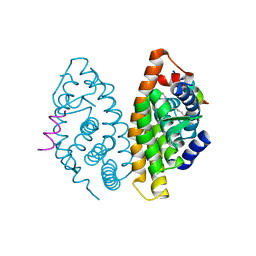 | | Crystal Structure of human retinoid X receptor alpha-ligand binding domain complex with 9cUAB76 and the coactivator peptide GRIP-1 | | Descriptor: | (3S,7S,8E)-8-[3-ethyl-2-(3-methylbutyl)cyclohex-2-en-1-ylidene]-3,7-dimethyloctanoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Xia, G, Muccio, D.D. | | Deposit date: | 2014-10-21 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformationally Defined Rexinoids and Their Efficacy in the Prevention of Mammary Cancers.
J.Med.Chem., 58, 2015
|
|
4RBV
 
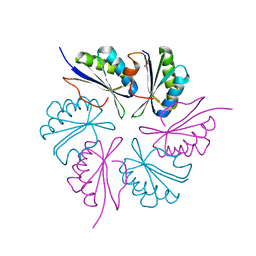 | |
3LQM
 
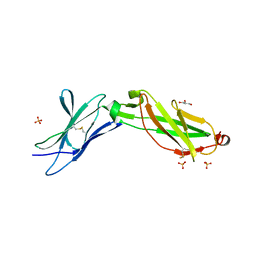 | | Structure of the IL-10R2 Common Chain | | Descriptor: | GLYCEROL, Interleukin-10 receptor subunit beta, SULFATE ION | | Authors: | Yoon, S.I, Walter, M.R. | | Deposit date: | 2010-02-09 | | Release date: | 2010-05-26 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure and mechanism of receptor sharing by the IL-10R2 common chain.
Structure, 18, 2010
|
|
3O52
 
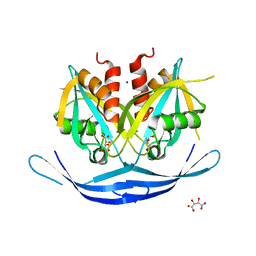 | | Structure of the E.coli GDP-mannose hydrolase (yffh) in complex with tartrate | | Descriptor: | CHLORIDE ION, D(-)-TARTARIC ACID, GDP-mannose pyrophosphatase nudK, ... | | Authors: | Amzel, L.M, Gabelli, S.B, Boto, A.N. | | Deposit date: | 2010-07-27 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural studies of the Nudix GDP-mannose hydrolase from E. coli reveals a new motif for mannose recognition.
Proteins, 79, 2011
|
|
3O6Z
 
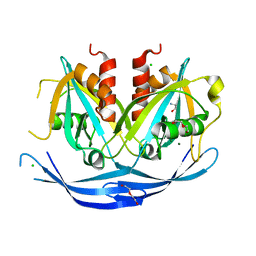 | | Structure of the D152A E.coli GDP-mannose hydrolase (yffh) in complex with Mg++ | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Amzel, L.M, Gabelli, S.B, Boto, A.N. | | Deposit date: | 2010-07-29 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural studies of the Nudix GDP-mannose hydrolase from E. coli reveals a new motif for mannose recognition.
Proteins, 79, 2011
|
|
3O61
 
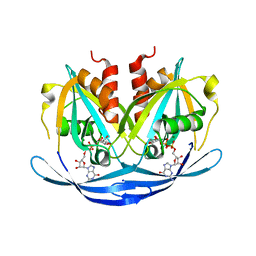 | | Structure of the E100A E.coli GDP-mannose hydrolase (yffh) in complex with GDP-mannose and Mg++ | | Descriptor: | CHLORIDE ION, GDP-mannose pyrophosphatase nudK, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, ... | | Authors: | Amzel, L.M, Gabelli, S.B, Boto, A.N. | | Deposit date: | 2010-07-28 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural studies of the Nudix GDP-mannose hydrolase from E. coli reveals a new motif for mannose recognition.
Proteins, 79, 2011
|
|
3O69
 
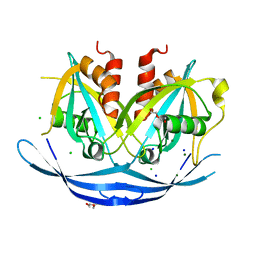 | | Structure of the E100A E.coli GDP-mannose hydrolase (yffh) in complex with Mg++ | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GDP-mannose pyrophosphatase nudK, ... | | Authors: | Amzel, L.M, Gabelli, S.B, Boto, A.N. | | Deposit date: | 2010-07-28 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural studies of the Nudix GDP-mannose hydrolase from E. coli reveals a new motif for mannose recognition.
Proteins, 79, 2011
|
|
6E7G
 
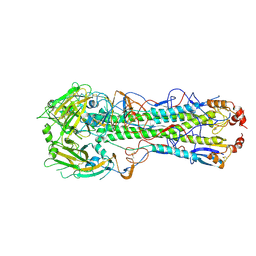 | |
6E7H
 
 | |
7TB0
 
 | | E. faecium MurAA in complex with fosfomycin and UNAG | | Descriptor: | CHLORIDE ION, POTASSIUM ION, SODIUM ION, ... | | Authors: | Zhou, Y, Shamoo, Y. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Enolpyruvate transferase MurAA A149E , identified during adaptation of Enterococcus faecium to daptomycin, increases stability of MurAA-MurG interaction.
J.Biol.Chem., 299, 2023
|
|
7TMA
 
 | |
7TME
 
 | |
7TLT
 
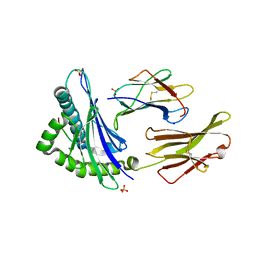 | | SARS-CoV-2 Spike-derived peptide S489-497 (YFPLQSYGF) presented by HLA-A*29:02 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A alpha chain, ... | | Authors: | Murdolo, L.D, Szeto, C, Gras, S. | | Deposit date: | 2022-01-18 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ablation of CD8 + T cell recognition of an immunodominant epitope in SARS-CoV-2 Omicron variants BA.1, BA.2 and BA.3.
Nat Commun, 13, 2022
|
|
