8BW0
 
 | | Structure of CEACAM5 A3-B3 domain in Complex with Tusamitamab Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Carcinoembryonic antigen-related cell adhesion molecule 5, ... | | Authors: | Kumar, A, Bertrand, T, Rapisarda, C, Rak, A. | | Deposit date: | 2022-12-06 | | Release date: | 2024-01-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural insights into epitope-paratope interactions of a monoclonal antibody targeting CEACAM5-expressing tumors.
Nat Commun, 15, 2024
|
|
8C4A
 
 | |
8BR4
 
 | | Structure of GAPDH from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kumar, A, Karthikeyan, S. | | Deposit date: | 2022-11-22 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Stoichiometry of ligand binding and role of C-terminal lysines in Mycobacterium tuberculosis and human GAPDH multifunctionality.
Febs J., 2024
|
|
4LQ6
 
 | | Crystal structure of Rv3717 reveals a novel amidase from M. tuberculosis | | Descriptor: | CHLORIDE ION, N-acetymuramyl-L-alanine amidase-related protein, PLATINUM (II) ION, ... | | Authors: | Kumar, A, Kumar, S, Kumar, D, Mishra, A, Dewangan, R.P, Shrivastava, P, Ramachandran, S, Taneja, B. | | Deposit date: | 2013-07-17 | | Release date: | 2013-12-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The structure of Rv3717 reveals a novel amidase from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3KJZ
 
 | | Crystal structure of native peptidyl-tRNA hydrolase from Mycobacterium smegmatis | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Kumar, A, Singh, N, Yadav, R, Prem Kumar, R, Sharma, S, Arora, A, Singh, T.P. | | Deposit date: | 2009-11-04 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of peptidyl-tRNA hydrolase from mycobacterium smegmatis reveals novel features related to enzyme dynamics.
Int J Biochem Mol Biol, 3, 2012
|
|
6JQV
 
 | | Crystal structure of Arabidopsis thaliana NRP2 | | Descriptor: | NAP1-related protein 2 | | Authors: | Kumar, A, Vasudevan, D. | | Deposit date: | 2019-04-01 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Structural Characterization ofArabidopsis thalianaNAP1-Related Protein 2 (AtNRP2) and Comparison with its Homolog AtNRP1.
Molecules, 24, 2019
|
|
5N74
 
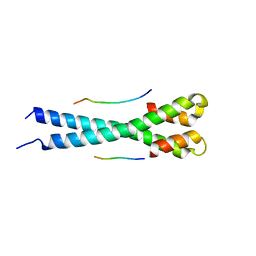 | | Microtubule end binding protein complex | | Descriptor: | Karyogamy protein KAR9, Microtubule-associated protein RP/EB family member 1 | | Authors: | Kumar, A, Steinmetz, M. | | Deposit date: | 2017-02-18 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Short Linear Sequence Motif LxxPTPh Targets Diverse Proteins to Growing Microtubule Ends.
Structure, 25, 2017
|
|
1JYM
 
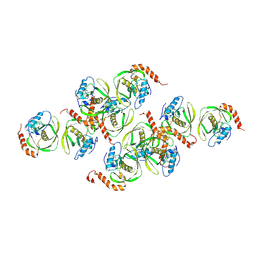 | | Crystals of Peptide Deformylase from Plasmodium falciparum with Ten Subunits per Asymmetric Unit Reveal Critical Characteristics of the Active Site for Drug Design | | Descriptor: | COBALT (II) ION, Peptide Deformylase | | Authors: | Kumar, A, Nguyen, K.T, Srivathsan, S, Ornstein, B, Turley, S, Hirsh, I, Pei, D, Hol, W.G.J. | | Deposit date: | 2001-09-12 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystals of peptide deformylase from Plasmodium falciparum reveal critical characteristics of the active site for drug design.
Structure, 10, 2002
|
|
5N2W
 
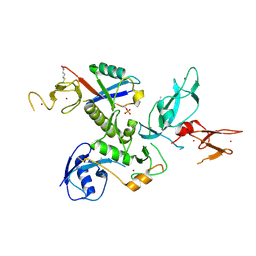 | | WT-Parkin and pUB complex | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase parkin,E3 ubiquitin-protein ligase parkin, Polyubiquitin-B, ... | | Authors: | Kumar, A, Chaugule, V.K, Johnson, C, Toth, R, Sundaramoorthy, R, Knebel, A, Walden, H. | | Deposit date: | 2017-02-08 | | Release date: | 2017-04-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Parkin-phosphoubiquitin complex reveals cryptic ubiquitin-binding site required for RBR ligase activity.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5N38
 
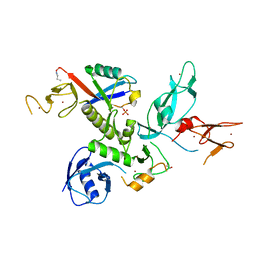 | | S65DParkin and pUB complex | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase parkin,E3 ubiquitin-protein ligase parkin, ... | | Authors: | Kumar, A, Chaugule, V.K, Johnson, C, Toth, R, Sundaramoorthy, R, Knebel, A, Walden, H. | | Deposit date: | 2017-02-08 | | Release date: | 2017-04-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Parkin-phosphoubiquitin complex reveals cryptic ubiquitin-binding site required for RBR ligase activity.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5OAT
 
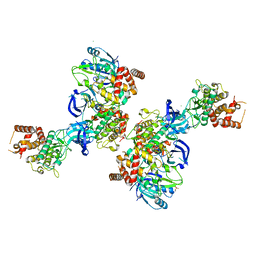 | | PINK1 structure | | Descriptor: | MAGNESIUM ION, Serine/threonine-protein kinase PINK1, mitochondrial-like Protein | | Authors: | Kumar, A, Tamjar, J, Woodroof, H.I, Raimi, O.G, Waddell, A.Y, Peggie, M, Muqit, M.M.K, van Aalten, D.M.F. | | Deposit date: | 2017-06-23 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structure of PINK1 and mechanisms of Parkinson's disease associated mutations.
Elife, 6, 2017
|
|
1CEH
 
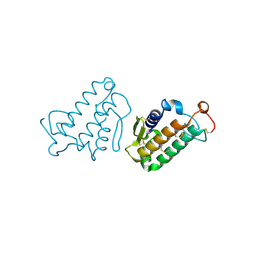 | | STRUCTURE AND FUNCTION OF THE CATALYTIC SITE MUTANT ASP99ASN OF PHOSPHOLIPASE A2: ABSENCE OF CONSERVED STRUCTURAL WATER | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Kumar, A, Sekharudu, C, Ramakrishnan, B, Dupureur, C.M, Zhu, H, Tsai, M.-D, Sundaralingam, M. | | Deposit date: | 1994-11-16 | | Release date: | 1995-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of the catalytic site mutant Asp 99 Asn of phospholipase A2: absence of the conserved structural water.
Protein Sci., 3, 1994
|
|
6Y90
 
 | |
6Y97
 
 | |
6Y92
 
 | |
6Y9A
 
 | |
7AG9
 
 | |
8IA6
 
 | |
8JCR
 
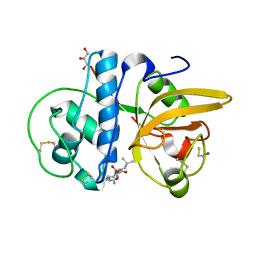 | | Crystal structure of Procerain from Calotropis gigantea (pH 6.0) | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE, ... | | Authors: | Kumar, A, Jamdar, S.N, Srivastava, G, Makde, R.D. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of Procerain from Calotropis gigantea
To Be Published
|
|
8JCQ
 
 | |
8JCS
 
 | |
8OH9
 
 | | Cryo-EM structure of the electron bifurcating transhydrogenase StnABC complex from Sporomusa Ovata (state 1) | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Formate dehydrogenase-O, ... | | Authors: | Kumar, A, Kremp, F, Mueller, V, Schuller, J.M. | | Deposit date: | 2023-03-20 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular architecture and electron transfer pathway of the Stn family transhydrogenase.
Nat Commun, 14, 2023
|
|
8OH5
 
 | | Cryo-EM structure of the electron bifurcating transhydrogenase StnABC complex from Sporomusa Ovata (state 2) | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kumar, A, Kremp, F, Mueller, V, Schuller, J.M. | | Deposit date: | 2023-03-20 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular architecture and electron transfer pathway of the Stn family transhydrogenase.
Nat Commun, 14, 2023
|
|
2RKW
 
 | | Intermediate position of ATP on its trail to the binding pocket inside the subunit B mutant R416W of the energy converter A1Ao ATP synthase | | Descriptor: | V-type ATP synthase beta chain | | Authors: | Kumar, A, Manimekalai, M.S.S, Balakrishna, A.M, Hunke, C, Gruber, G. | | Deposit date: | 2007-10-18 | | Release date: | 2008-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Spectroscopic and crystallographic studies of the mutant R416W give insight into the nucleotide binding traits of subunit B of the A1Ao ATP synthase
Proteins, 75, 2009
|
|
6JMI
 
 | | Crystal structure of M.tuberculosis Rv0081 | | Descriptor: | SULFATE ION, Uncharacterized HTH-type transcriptional regulator Rv0081 | | Authors: | Kumar, A, Phulera, S, Mande, C.S. | | Deposit date: | 2019-03-11 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.896 Å) | | Cite: | Structural basis of hypoxic gene regulation by the Rv0081 transcription factor of Mycobacterium tuberculosis.
Febs Lett., 593, 2019
|
|
