6QRZ
 
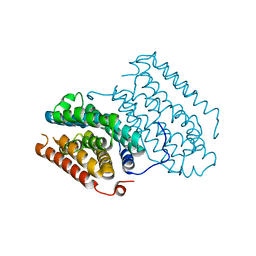 | | Crystal structure of R2-like ligand-binding oxidase from Sulfolobus acidocaldarius solved by 3D micro-crystal electron diffraction | | Descriptor: | FE (III) ION, MANGANESE (III) ION, Ribonucleoside-diphosphate reductase | | Authors: | Xu, H, Lebrette, H, Clabbers, M.T.B, Zhao, J, Griese, J.J, Zou, X, Hogbom, M. | | Deposit date: | 2019-02-20 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON CRYSTALLOGRAPHY (3 Å) | | Cite: | Solving a new R2lox protein structure by microcrystal electron diffraction.
Sci Adv, 5, 2019
|
|
7MYO
 
 | | Cryo-EM structure of p110alpha in complex with p85alpha inhibited by BYL-719 | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Yang, S, Hart, J.R, Xu, Y, Zou, X, Zhang, H, Zhou, Q, Xia, T, Zhang, Y, Yang, D, Wang, M.-W, Vogt, P.K. | | Deposit date: | 2021-05-21 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM structures of PI3K alpha reveal conformational changes during inhibition and activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MYN
 
 | | Cryo-EM Structure of p110alpha in complex with p85alpha | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Yang, S, Hart, J.R, Xu, Y, Zou, X, Zhang, H, Zhou, Q, Xia, T, Zhang, Y, Yang, D, Wang, M.-W, Vogt, P.K. | | Deposit date: | 2021-05-21 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Cryo-EM structures of PI3K alpha reveal conformational changes during inhibition and activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6YMB
 
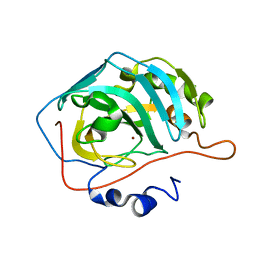 | | MicroED structure of human carbonic anhydrase II | | Descriptor: | ZINC ION, carbonic anhydrase 2 | | Authors: | Clabbers, M.T.B, Fisher, S.Z, Coincon, M, Zou, X, Xu, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.5 Å) | | Cite: | Visualizing drug binding interactions using microcrystal electron diffraction.
Commun Biol, 3, 2020
|
|
6YMA
 
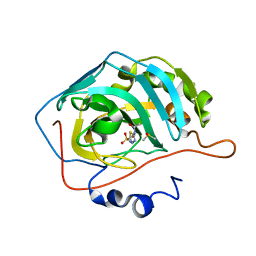 | | MicroED structure of acetazolamide-bound human carbonic anhydrase II | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, DIMETHYL SULFOXIDE, ZINC ION, ... | | Authors: | Clabbers, M.T.B, Fisher, S.Z, Coincon, M, Zou, X, Xu, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.5 Å) | | Cite: | Visualizing drug binding interactions using microcrystal electron diffraction.
Commun Biol, 3, 2020
|
|
5OCV
 
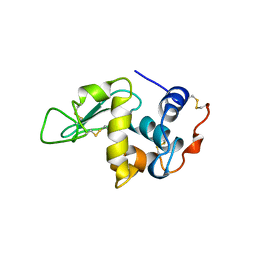 | | A Rare Lysozyme Crystal Form Solved Using High-Redundancy 3D Electron Diffraction Data from Micron-Sized Needle Shaped Crystals | | Descriptor: | Lysozyme C, SODIUM ION | | Authors: | Xu, H, Lebrette, H, Yang, T, Srinivas, V, Hovmoller, S, Hogbom, M, Zou, X. | | Deposit date: | 2017-07-03 | | Release date: | 2018-03-28 | | Last modified: | 2024-01-17 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.2 Å) | | Cite: | A Rare Lysozyme Crystal Form Solved Using Highly Redundant Multiple Electron Diffraction Datasets from Micron-Sized Crystals.
Structure, 26, 2018
|
|
8DCP
 
 | | PI 3-kinase alpha with nanobody 3-126 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DD4
 
 | | PI 3-kinase alpha with nanobody 3-142 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DD8
 
 | | PI 3-kinase alpha with nanobody 3-142, crosslinked with DSG | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DCX
 
 | | PI 3-kinase alpha with nanobody 3-159 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4DN8
 
 | | Structure of porcine surfactant protein D neck and carbohydrate recognition domain complexed with mannose | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein D, beta-D-mannopyranose | | Authors: | van Eijk, M, Rynkiewicz, M.J, White, M.R, Hartshorn, K.L, Zou, X, Schulten, K, Luo, D, Crouch, E.C, Cafarella, T.M, Head, J.F, Haagsman, H.P, Seaton, B.A. | | Deposit date: | 2012-02-08 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Unique Sugar-binding Site Mediates the Distinct Anti-influenza Activity of Pig Surfactant Protein D.
J.Biol.Chem., 287, 2012
|
|
7DUQ
 
 | | Cryo-EM structure of the compound 2 and GLP-1-bound human GLP-1 receptor-Gs complex | | Descriptor: | CHOLESTEROL, Glucagon-like peptide 1, Glucagon-like peptide 1 receptor, ... | | Authors: | Cong, Z, Chen, L, Ma, H, Zhou, Q, Zou, X, Ye, C, Dai, A, Liu, Q, Huang, W, Sun, X, Wang, X, Xu, P, Zhao, L, Xia, T, Zhong, W, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2021-01-11 | | Release date: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular insights into ago-allosteric modulation of the human glucagon-like peptide-1 receptor.
Nat Commun, 12, 2021
|
|
7DUR
 
 | | Cryo-EM structure of the compound 2-bound human GLP-1 receptor-Gs complex | | Descriptor: | CHOLESTEROL, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cong, Z, Chen, L, Ma, H, Zhou, Q, Zou, X, Ye, C, Dai, A, Liu, Q, Huang, W, Sun, X, Wang, X, Xu, P, Zhao, L, Xia, T, Zhong, W, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2021-01-11 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular insights into ago-allosteric modulation of the human glucagon-like peptide-1 receptor.
Nat Commun, 12, 2021
|
|
7EVM
 
 | | Cryo-EM structure of the compound 2-bound human GLP-1 receptor-Gs complex | | Descriptor: | CHOLESTEROL, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cong, Z, Chen, L, Ma, H, Zhou, Q, Zou, X, Ye, C, Dai, A, Liu, Q, Huang, W, Sun, X, Wang, X, Xu, P, Zhao, L, Xia, T, Zhong, W, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2021-05-21 | | Release date: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular insights into ago-allosteric modulation of the human glucagon-like peptide-1 receptor.
Nat Commun, 12, 2021
|
|
7F16
 
 | | Cryo-EM structure of parathyroid hormone receptor type 2 in complex with a tuberoinfundibular peptide of 39 residues and G protein | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, X, Cheng, X, Zhao, L.H, Wang, Y, Ye, C, Zou, X, Dai, A, Cong, Z, Chen, J, Zhou, Q, Xia, T, Jiang, H, Xu, H.E, Yang, D, Wang, M.W. | | Deposit date: | 2021-06-08 | | Release date: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular insights into differentiated ligand recognition of the human parathyroid hormone receptor 2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LVA
 
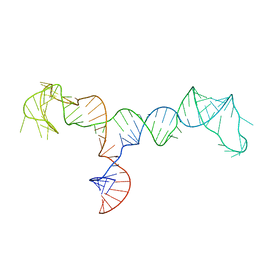 | | Solution structure of the HIV-1 PBS-segment | | Descriptor: | RNA (103-MER) | | Authors: | Heng, X, Song, Z. | | Deposit date: | 2021-02-24 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | The three-way junction structure of the HIV-1 PBS-segment binds host enzyme important for viral infectivity.
Nucleic Acids Res., 49, 2021
|
|
8Y2T
 
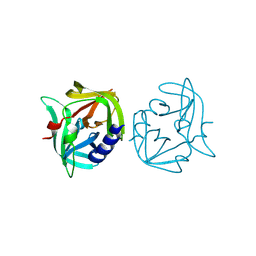 | | Crystal structure of 3C protease from coxsackievirus B3 | | Descriptor: | Protease 3C | | Authors: | Jiang, H.H, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2024-01-27 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the 3C proteases from Coxsackievirus B3 and B4.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
8WZQ
 
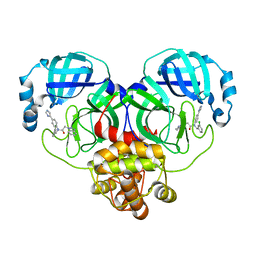 | | Crystal structure of SARS-Cov-2 main protease V186F mutant in complex with CCF0058981 | | Descriptor: | 2-(benzotriazol-1-yl)-~{N}-[(3-chlorophenyl)methyl]-~{N}-[4-(1~{H}-imidazol-5-yl)phenyl]ethanamide, 3C-like proteinase nsp5 | | Authors: | Zou, X.F, Jiang, H.H, Zhou, X.L, Zhang, J, Li, J. | | Deposit date: | 2023-11-02 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease (M pro ) mutants in complex with the non-covalent inhibitor CCF0058981.
Biochem.Biophys.Res.Commun., 692, 2024
|
|
8WZP
 
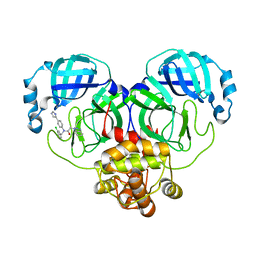 | | Crystal structure of SARS-Cov-2 main protease M49I mutant in complex with CCF0058981 | | Descriptor: | 2-(benzotriazol-1-yl)-~{N}-[(3-chlorophenyl)methyl]-~{N}-[4-(1~{H}-imidazol-5-yl)phenyl]ethanamide, 3C-like proteinase nsp5 | | Authors: | Jiang, H.H, Zou, X.F, Zhou, X.L, Zhang, J, Li, J. | | Deposit date: | 2023-11-02 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease (M pro ) mutants in complex with the non-covalent inhibitor CCF0058981.
Biochem.Biophys.Res.Commun., 692, 2024
|
|
8HVZ
 
 | | Crystal structure of SARS-Cov-2 main protease V186F mutant in complex with PF07304814 | | Descriptor: | 3C-like proteinase nsp5, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2022-12-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of main protease (M pro ) mutants of SARS-CoV-2 variants bound to PF-07304814.
Mol Biomed, 4, 2023
|
|
8HVW
 
 | | Crystal structure of SARS-Cov-2 main protease M49I mutant in complex with PF07304814 | | Descriptor: | 3C-like proteinase nsp5, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2022-12-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of main protease (M pro ) mutants of SARS-CoV-2 variants bound to PF-07304814.
Mol Biomed, 4, 2023
|
|
8IG9
 
 | |
7V9M
 
 | | Cryo-EM structure of the GHRH-bound human GHRHR splice variant 1 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Cong, Z.T, Zhou, F.L, Zhang, C, Zou, X.Y, Zhang, H.B, Wang, Y.Z, Zhou, Q.T, Cai, X.Q, Liu, Q.F, Li, J, Shao, L.J, Mao, C.Y, Wang, X, Wu, J.H, Xia, T, Zhao, L.H, Jiang, H.L, Zhang, Y, Xu, H.E, Chen, X, Yang, D.H, Wang, M.W. | | Deposit date: | 2021-08-26 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Constitutive signal bias mediated by the human GHRHR splice variant 1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7V9L
 
 | | Cryo-EM structure of the SV1-Gs complex. | | Descriptor: | GHRH receptor splice variant 1,GHRH receptor splice variant 1,GHRH receptor splice variant 1,SV1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cong, Z.T, Zhou, F.L, Zhang, C, Zou, X.Y, Zhang, H.B, Wang, Y.Z, Zhou, Q.T, Cai, X.Q, Liu, Q.F, Li, J, Shao, L.J, Mao, C.Y, Wang, X, Wu, J.H, Xia, T, Zhao, L.H, Jiang, H.L, Zhang, Y, Xu, H.E, Cheng, X, Yang, D.H, Wang, M.W. | | Deposit date: | 2021-08-26 | | Release date: | 2022-04-06 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Constitutive signal bias mediated by the human GHRHR splice variant 1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6RTI
 
 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with aptamer A9g | | Descriptor: | (2S)-2-(PHOSPHONOMETHYL)PENTANEDIOIC ACID, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Motlova, L, Kolenko, P, Barinka, C. | | Deposit date: | 2019-05-24 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of prostate-specific membrane antigen recognition by the A9g RNA aptamer.
Nucleic Acids Res., 48, 2020
|
|
