8E1F
 
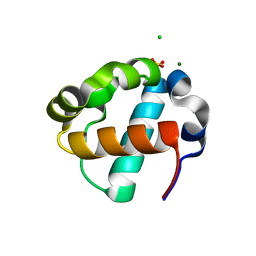 | | Sterile Alpha Motif Domain of Human Translocation ETS Leukemia, Non-Polymer Crystal Form | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Wilson, E, Nawarathnage, S, Stewart, C, Brown, S, Bezzant, B, Bunn, D, Towne, N, Pedroza Romo, M.J, Moody, J.D. | | Deposit date: | 2022-08-10 | | Release date: | 2022-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Sterile Alpha Motif Domain of Human Translocation ETS Leukemia, Non-Polymer Crystal Form
To Be Published
|
|
7N2B
 
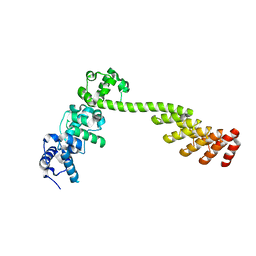 | | A DARPin semi-rigidly fused to the 3TEL crystallization chaperone | | Descriptor: | Transcription factor ETV6,Transcription factor ETV6,Transcription factor ETV6,3TEL-rigid-DARPin | | Authors: | Sarath Nawarathange, S.D, Gajjar, P, Bunn, D, Stewart, C, Doukov, T, Moody, J.D. | | Deposit date: | 2021-05-28 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.221 Å) | | Cite: | Crystals of TELSAM-target protein fusions that exhibit minimal crystal contacts and lack direct inter-TELSAM contacts.
Open Biology, 12, 2022
|
|
7TDY
 
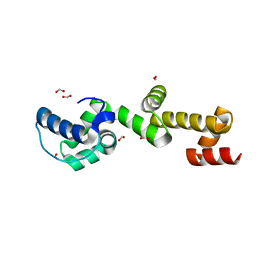 | | The ubiquitin-associated domain of human thirty-eight negative kinase 1, flexibly fused to the 1TEL crystallization chaperone via a 2-glycine linker and crystallized at low protein concentration | | Descriptor: | FORMIC ACID, Transcription factor ETV6,Non-receptor tyrosine-protein kinase TNK1 | | Authors: | Nawarathnage, S, Bunn, D.R, Stewart, C, Doukev, T, Moody, J.D. | | Deposit date: | 2022-01-03 | | Release date: | 2023-01-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Fusion crystallization reveals the behavior of both the 1TEL crystallization chaperone and the TNK1 UBA domain.
Structure, 31, 2023
|
|
7TCY
 
 | | The ubiquitin-associated domain of human thirty-eight negative kinase I | | Descriptor: | CHLORIDE ION, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Nawarathnage, S, Bunn, R.D, Stewart, C, Doukov, T, Moody, J.D. | | Deposit date: | 2021-12-29 | | Release date: | 2023-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Fusion crystallization reveals the behavior of both the 1TEL crystallization chaperone and the TNK1 UBA domain.
Structure, 31, 2023
|
|
7U4W
 
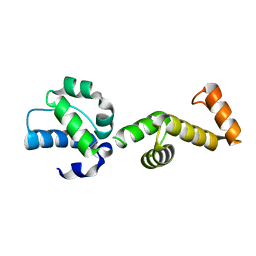 | | The ubiquitin-associated domain of human thirty-eight negative kinase-1 flexibly fused to the 1TEL crystallization chaperone via a 2-glycine linker and crystallized at traditional protein concentration | | Descriptor: | Transcription factor ETV6,Non-receptor tyrosine-protein kinase TNK1 | | Authors: | Nawarathnage, S, Pedroza Romo, M.J, Smith, T, Bunn, D, Stewart, C, Doukov, T, Moody, J.D. | | Deposit date: | 2022-03-01 | | Release date: | 2023-03-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fusion crystallization reveals the behavior of both the 1TEL crystallization chaperone and the TNK1 UBA domain.
Structure, 31, 2023
|
|
5W8Q
 
 | |
5WC4
 
 | |
5UCO
 
 | |
5UC5
 
 | | Chalcone synthase from Malus domestica | | Descriptor: | CHS2 chalcone synthase | | Authors: | Stewart Jr, C.E, Noel, J.P. | | Deposit date: | 2016-12-21 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Molecular architectures of benzoic acid-specific type III polyketide synthases.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
8S9G
 
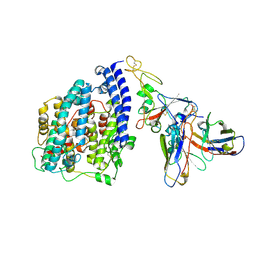 | | SARS-CoV-2 BN.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, S309 Fab Heavy chain, ... | | Authors: | Park, Y.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2023-03-28 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Neutralization, effector function and immune imprinting of Omicron variants.
Nature, 621, 2023
|
|
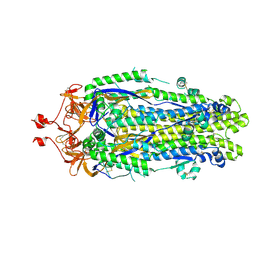
8VQ9
 
 | |
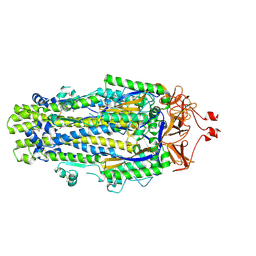
8VQB
 
 | |
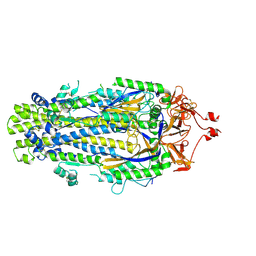
8VQA
 
 | |
8FT8
 
 | | The von Willebrand factor A domain of human capillary morphogenesis gene II, flexibly fused to the 1TEL crystallization chaperone, Thr-Val linker variant, SUMO tag-free preparation | | Descriptor: | CHLORIDE ION, GLYCEROL, POTASSIUM ION, ... | | Authors: | Gajjar, P.L, Litchfield, C.M, Callahan, M, Redd, N, Doukov, T, Moody, J.D. | | Deposit date: | 2023-01-11 | | Release date: | 2023-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Increasing the bulk of the 1TEL-target linker and retaining the 10×His tag in a 1TEL-CMG2-vWa construct improves crystal order and diffraction limits.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8FZU
 
 | | The von Willebrand factor A domain of human capillary morphogenesis gene II, flexibly fused to the 1TEL crystallization chaperone, Thr-Val linker variant, Expressed with SUMO tag | | Descriptor: | POTASSIUM ION, SULFATE ION, Transcription factor ETV6,Anthrax toxin receptor 2 | | Authors: | Gajjar, P.L, Litchfield, C.M, Callahan, M, Redd, N, Doukov, T, Lebedev, A, Moody, J.D. | | Deposit date: | 2023-01-30 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Increasing the bulk of the 1TEL-target linker and retaining the 10×His tag in a 1TEL-CMG2-vWa construct improves crystal order and diffraction limits.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8FZV
 
 | | The von Willebrand factor A domain of human capillary morphogenesis gene II, flexibly fused to the 1TEL crystallization chaperone, Ala-Ala linker variant, expressed with SUMO tag | | Descriptor: | MAGNESIUM ION, Transcription factor ETV6,Anthrax toxin receptor 2, UNKNOWN ATOM OR ION | | Authors: | Pedroza Romo, M.J, Soleimani, S, Doukov, T, Lebedev, A, Moody, J.D. | | Deposit date: | 2023-01-30 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Increasing the bulk of the 1TEL-target linker and retaining the 10×His tag in a 1TEL-CMG2-vWa construct improves crystal order and diffraction limits.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8FT6
 
 | | The von Willebrand factor A domain of human capillary morphogenesis gene II, flexibly fused to the 1TEL crystallization chaperone, Ala-Ala linker variant, SUMO tag-free preparation. | | Descriptor: | CITRIC ACID, IODIDE ION, SULFATE ION, ... | | Authors: | Gajjar, P.L, Litchfield, C.M, Callahan, M, Redd, N, Doukov, T, Moody, J.D. | | Deposit date: | 2023-01-11 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Increasing the bulk of the 1TEL-target linker and retaining the 10×His tag in a 1TEL-CMG2-vWa construct improves crystal order and diffraction limits.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8FXB
 
 | | SARS-CoV-2 XBB.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, S309 Heavy chain, ... | | Authors: | Park, Y.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2023-01-24 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Neutralization, effector function and immune imprinting of Omicron variants.
Nature, 621, 2023
|
|
8FXC
 
 | | SARS-CoV-2 BQ.1.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, S309 Heavy chain, ... | | Authors: | Park, Y.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2023-01-24 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Neutralization, effector function and immune imprinting of Omicron variants.
Nature, 621, 2023
|
|
8ERQ
 
 | |
8ERR
 
 | |
8U0T
 
 | |
8U29
 
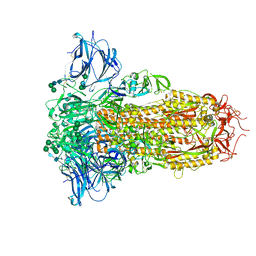 | | Prefusion structure of the PRD-0038 spike glycoprotein ectodomain trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PRD-0038 Spike glycoprotein, ... | | Authors: | Lee, J, Park, Y.J, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2023-09-05 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Broad receptor tropism and immunogenicity of a clade 3 sarbecovirus.
Cell Host Microbe, 31, 2023
|
|
8VGT
 
 | |
8DYA
 
 | |
