4KU1
 
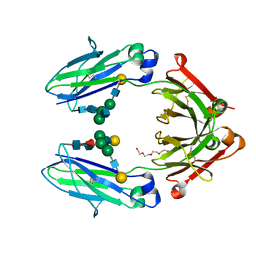 | | Role of the hinge and C-gamma-2/C-gamma-3 interface in immunoglobin G1 Fc domain motions: implications for Fc engineering | | Descriptor: | Ig gamma-1 chain C region, TETRAETHYLENE GLYCOL, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Frank, M, Walker, R, Lanzilotta, W.N, Prestegard, J.H, Barb, A.W. | | Deposit date: | 2013-05-21 | | Release date: | 2014-02-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Immunoglobulin g1 fc domain motions: implications for fc engineering.
J.Mol.Biol., 426, 2014
|
|
1XVL
 
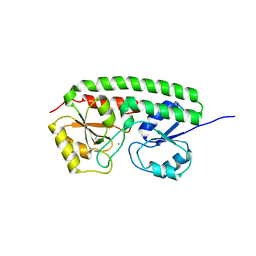 | | The three-dimensional structure of MntC from Synechocystis 6803 | | Descriptor: | MANGANESE (II) ION, Mn transporter | | Authors: | Rukhman, V, Anati, R, Melamed-Frank, M, Bhattacharyya-Pakrasi, M, Pakrasi, H.B, Adir, N. | | Deposit date: | 2004-10-28 | | Release date: | 2005-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The MntC crystal structure suggests that import of Mn2+ in cyanobacteria is redox controlled.
J.Mol.Biol., 348, 2005
|
|
3RTK
 
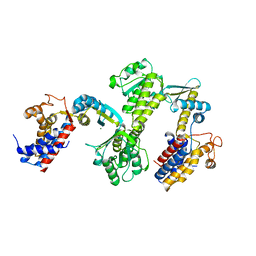 | | Crystal structure of Cpn60.2 from Mycobacterium tuberculosis at 2.8A | | Descriptor: | 60 kDa chaperonin 2, MAGNESIUM ION | | Authors: | Shahar, A, Melamed-Frank, M, Kashi, Y, Adir, N. | | Deposit date: | 2011-05-03 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The dimeric structure of the Cpn60.2 chaperonin of Mycobacterium tuberculosis at 2.8 A reveals possible modes of function.
J.Mol.Biol., 412, 2011
|
|
4RVY
 
 | | Serial Time resolved crystallography of Photosystem II using a femtosecond X-ray laser. The S state after two flashes (S3) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kupitz, C, Basu, S, Grotjohann, I, Fromme, R, Zatsepin, N, Rendek, K.N, Hunter, M, Shoeman, R.L, White, T.A, Wang, D, James, D, Yang, J.-H, Cobb, D.E, Reeder, B, Sierra, R.G, Liu, H, Barty, A, Aquila, A, Deponte, D, Kirian, R, Bari, S, Bergkamp, J.J, Beyerlein, K, Bogan, M.J, Caleman, C, Chao, T.-C, Conrad, C.E, Davis, K.M, Fleckenstein, H, Galli, L, Hau-Riege, S.P, Kassemeyer, S, Laksmono, H, Liang, M, Lomb, L, Marchesini, S, Martin, A.V, Messerschmidt, M, Milathianaki, D, Nass, K, Ros, A, Roy-Chowdhury, S, Schmidt, K, Seibert, M, Steinbrener, J, Stellato, F, Yan, L, Yoon, C, Moore, T.A, Moore, A.L, Pushkar, Y, Williams, G.J, Boutet, S, Doak, R.B, Weierstall, U, Frank, M, Chapman, H.N, Spence, J.C.H, Fromme, P. | | Deposit date: | 2014-11-29 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Serial time-resolved crystallography of photosystem II using a femtosecond X-ray laser.
Nature, 513, 2014
|
|
7Q64
 
 | | Cryo-em structure of the Nup98 fibril polymorph 1 | | Descriptor: | Nuclear pore complex protein Nup98 | | Authors: | Ibanez de Opakua, A, Geraets, J.A, Frieg, B, Dienemann, C, Savastano, A, Rankovic, M, Cima-Omori, M.-S, Schroeder, G.F, Zweckstetter, M. | | Deposit date: | 2021-11-05 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Molecular interactions of FG nucleoporin repeats at high resolution.
Nat.Chem., 14, 2022
|
|
7Q67
 
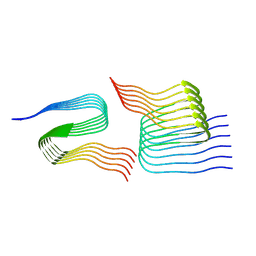 | | Cryo-em structure of the Nup98 fibril polymorph 4 | | Descriptor: | Nuclear pore complex protein Nup98 | | Authors: | Ibanez de Opakua, A, Geraets, J.A, Frieg, B, Dienemann, C, Savastano, A, Rankovic, M, Cima-Omori, M.-S, Schroeder, G.F, Zweckstetter, M. | | Deposit date: | 2021-11-05 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Molecular interactions of FG nucleoporin repeats at high resolution.
Nat.Chem., 14, 2022
|
|
7Q65
 
 | | Cryo-em structure of the Nup98 fibril polymorph 2 | | Descriptor: | Nuclear pore complex protein Nup98 | | Authors: | Ibanez de Opakua, A, Geraets, J.A, Frieg, B, Dienemann, C, Savastano, A, Rankovic, M, Cima-Omori, M.-S, Schroeder, G.F, Zweckstetter, M. | | Deposit date: | 2021-11-05 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Molecular interactions of FG nucleoporin repeats at high resolution.
Nat.Chem., 14, 2022
|
|
7Q66
 
 | | Cryo-em structure of the Nup98 fibril polymorph 3 | | Descriptor: | Nuclear pore complex protein Nup98 | | Authors: | Ibanez de Opakua, A, Geraets, J.A, Frieg, B, Dienemann, C, Savastano, A, Rankovic, M, Cima-Omori, M.-S, Schroeder, G.F, Zweckstetter, M. | | Deposit date: | 2021-11-05 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Molecular interactions of FG nucleoporin repeats at high resolution.
Nat.Chem., 14, 2022
|
|
5J7A
 
 | | Bacteriorhodopsin ground state structure obtained with Serial Femtosecond Crystallography | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Nogly, P, Panneels, V, Nelson, G, Gati, C, Kimura, T, Milne, C, Milathianaki, D, Kubo, M, Wu, W, Conrad, C, Coe, J, Bean, R, Zhao, Y, Bath, P, Dods, R, Harimoorthy, R, Beyerlein, K.R, Rheinberger, J, James, D, DePonte, D, Li, C, Sala, L, Williams, G, Hunter, M, Koglin, J.E, Berntsen, P, Nango, E, Iwata, S, Chapman, H.N, Fromme, P, Frank, M, Abela, R, Boutet, S, Barty, A, White, T.A, Weierstall, U, Spence, J, Neutze, R, Schertler, G, Standfuss, J. | | Deposit date: | 2016-04-06 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lipidic cubic phase injector is a viable crystal delivery system for time-resolved serial crystallography.
Nat Commun, 7, 2016
|
|
7TN9
 
 | | Structure of the Inmazeb cocktail and resistance to escape against Ebola virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein, GP2, ... | | Authors: | Rayaprolu, V, Fulton, B, Rafique, A, Arturo, E, Williams, D, Hariharan, C, Callaway, H, Parvate, A, Schendel, S.L, Parekh, D, Hui, S, Shaffer, K, Pascal, K.E, Wloga, E, Giordano, S, Copin, R, Franklin, M, Boytz, R.M, Donahue, C, Davey, R, Baum, A, Kyratsous, C.A, Saphire, E.O. | | Deposit date: | 2022-01-20 | | Release date: | 2023-01-25 | | Last modified: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the Inmazeb cocktail and resistance to Ebola virus escape.
Cell Host Microbe, 31, 2023
|
|
4PBU
 
 | | Serial Time-resolved crystallography of Photosystem II using a femtosecond X-ray laser The S1 state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Kupitz, C, Basu, S, Grotjohann, I, Fromme, R, Zatsepin, N, Rendek, K.N, Hunter, M, Shoeman, R.L, White, T.A, Wang, D, James, D, Yang, J.H, Cobb, D.E, Reeder, B, Sierra, R.G, Liu, H, Barty, A, Aquila, A, Deponte, D, Kirian, R.A, Bari, S, Bergkamp, J.J, Beyerlein, K, Bogan, M.J, Caleman, C, Chao, T.-C, Conrad, C.E, Davis, K.M, Fleckenstein, H, Galli, L, Hau-Riege, S.P, Kassemeyer, S, Laksmono, H, Liang, M, Lomb, L, Marchesini, S, Martin, A.V, Messerschmidt, M, Milathianaki, D, Nass, K, Ros, A, Roy-Chowdhury, S, Schmidt, K, Seibert, M, Steinbrener, J, Stellato, F, Yan, L, Yoon, C, Moore, T.A, Moore, A.L, Pushkar, Y, Williams, G.J, Boutet, S, Doak, R.B, Weierstall, U, Frank, M, Chapman, H.N, Spence, J.C.H, Fromme, P. | | Deposit date: | 2014-04-13 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Serial time-resolved crystallography of photosystem II using a femtosecond X-ray laser.
Nature, 513, 2014
|
|
3PCQ
 
 | | Femtosecond X-ray protein Nanocrystallography | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Chapman, H.N, Fromme, P, Barty, A, White, T.A, Kirian, R.A, Aquila, A, Hunter, M.S, Schulz, J, Deponte, D.P, Weierstall, U, Doak, R.B, Maia, F.R.N.C, Martin, A.V, Schlichting, I, Lomb, L, Coppola, N, Shoeman, R.L, Epp, S.W, Hartmann, R, Rolles, D, Rudenko, A, Foucar, L, Kimmel, N, Weidenspointner, G, Holl, P, Liang, M, Barthelmess, M, Caleman, C, Boutet, S, Bogan, M.J, Krzywinski, J, Bostedt, C, Bajt, S, Gumprecht, L, Rudek, B, Erk, B, Schmidt, C, Homke, A, Reich, C, Pietschner, D, Struder, L, Hauser, G, Gorke, H, Ullrich, J, Herrmann, S, Schaller, G, Schopper, F, Soltau, H, Kuhnel, K.-U, Messerschmidt, M, Bozek, J.D, Hau-Riege, S.P, Frank, M, Hampton, C.Y, Sierra, R, Starodub, D, Williams, G.J, Hajdu, J, Timneanu, N, Seibert, M.M, Andreasson, J, Rocker, A, Jonsson, O, Svenda, M, Stern, S, Nass, K, Andritschke, R, Schroter, C.-D, Krasniqi, F, Bott, M, Schmidt, K.E, Wang, X, Grotjohann, I, Holton, J.M, Barends, T.R.M, Neutze, R, Marchesini, S, Fromme, R, Schorb, S, Rupp, D, Adolph, M, Gorkhover, T, Andersson, I, Hirsemann, H, Potdevin, G, Graafsma, H, Nilsson, B, Spence, J.C.H. | | Deposit date: | 2010-10-21 | | Release date: | 2011-02-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (8.984 Å) | | Cite: | Femtosecond X-ray protein nanocrystallography.
Nature, 470, 2011
|
|
4CAS
 
 | | Serial femtosecond crystallography structure of a photosynthetic reaction center | | Descriptor: | (2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL TRIHYDROGEN DIPHOSPHATE, 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL A, ... | | Authors: | Johansson, L.C, Arnlund, D, Katona, G, White, T.A, Barty, A, DePonte, D.P, Shoeman, R.L, Wickstrand, C, Sharma, A, Williams, G.J, Aquila, A, Bogan, M.J, Caleman, C, Davidsson, J, Doak, R.B, Frank, M, Fromme, R, Galli, L, Grotjohann, I, Hunter, M.S, Kassemeyer, S, Kirian, R.A, Kupitz, C, Liang, M, Lomb, L, Malmerberg, E, Martin, A.V, Messerschmidt, M, Nass, K, Redecke, L, Seibert, M.M, Sjohamn, J, Steinbrener, J, Stellato, F, Wang, D, Wahlgren, W.Y, Weierstall, U, Westenhoff, S, Zatsepin, N.A, Boutet, S, Spence, J.C.H, Schlichting, I, Chapman, H.N, Fromme, P, Neutze, R. | | Deposit date: | 2013-10-09 | | Release date: | 2013-12-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of a photosynthetic reaction centre determined by serial femtosecond crystallography.
Nat Commun, 4, 2013
|
|
8FZW
 
 | | Thaumatin crystallized in cyclic olefin copolymer-based microfluidic chips | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Liu, Z, Gu, K, Shelby, M.L, Gilbile, D, Lyubimov, A.Y, Russi, S, Cohen, A.E, Coleman, M.A, Frank, M, Kuhl, T.L. | | Deposit date: | 2023-01-30 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | A user-friendly plug-and-play cyclic olefin copolymer-based microfluidic chip for room-temperature, fixed-target serial crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
4HWY
 
 | | Trypanosoma brucei procathepsin B solved from 40 fs free-electron laser pulse data by serial femtosecond X-ray crystallography | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cysteine peptidase C (CPC), beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Redecke, L, Nass, K, DePonte, D.P, White, T.A, Rehders, D, Barty, A, Stellato, F, Liang, M, Barends, T.R.M, Boutet, S, Williams, G.W, Messerschmidt, M, Seibert, M.M, Aquila, A, Arnlund, D, Bajt, S, Barth, T, Bogan, M.J, Caleman, C, Chao, T.-C, Doak, R.B, Fleckenstein, H, Frank, M, Fromme, R, Galli, L, Grotjohann, I, Hunter, M.S, Johansson, L.C, Kassemeyer, S, Katona, G, Kirian, R.A, Koopmann, R, Kupitz, C, Lomb, L, Martin, A.V, Mogk, S, Neutze, R, Shoemann, R.L, Steinbrener, J, Timneanu, N, Wang, D, Weierstall, U, Zatsepin, N.A, Spence, J.C.H, Fromme, P, Schlichting, I, Duszenko, M, Betzel, C, Chapman, H. | | Deposit date: | 2012-11-09 | | Release date: | 2012-12-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Natively inhibited Trypanosoma brucei cathepsin B structure determined by using an X-ray laser.
Science, 339, 2013
|
|
6E0I
 
 | | Crystal structure of Glucokinase in complex with compound 72 | | Descriptor: | 1-{4-[5-({3-[(2-methylpyridin-3-yl)oxy]-5-[(pyridin-2-yl)sulfanyl]pyridin-2-yl}amino)-1,2,4-thiadiazol-3-yl]piperidin-1 -yl}ethan-1-one, DIMETHYL SULFOXIDE, Glucokinase, ... | | Authors: | Hinklin, R.J, Baer, B.R, Boyd, S.A, Chicarelli, M.D, Condroski, K.R, DeWolf, W.E, Fischer, J, Frank, M, Hingorani, G.P, Lee, P.A, Neitzel, N.A, Pratt, S.A, Singh, A, Sullivan, F.X, Turner, T, Voegtli, W.C, Wallace, E.M, Williams, L, Aicher, T.D. | | Deposit date: | 2018-07-06 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and preclinical development of AR453588 as an anti-diabetic glucokinase activator.
Bioorg.Med.Chem., 28, 2020
|
|
6E0E
 
 | | Crystal structure of Glucokinase in complex with compound 6 | | Descriptor: | 2-({2-[(4-methyl-1,3-thiazol-2-yl)amino]pyridin-3-yl}oxy)benzonitrile, Glucokinase, alpha-D-glucopyranose | | Authors: | Hinklin, R.J, Baer, B.R, Boyd, S.A, Chicarelli, M.D, Condroski, K.R, DeWolf, W.E, Fischer, J, Frank, M, Hingorani, G.P, Lee, P.A, Neitzel, N.A, Pratt, S.A, Singh, A, Sullivan, F.X, Turner, T, Voegtli, W.C, Wallace, E.M, Williams, L, Aicher, T.D. | | Deposit date: | 2018-07-06 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and preclinical development of AR453588 as an anti-diabetic glucokinase activator.
Bioorg.Med.Chem., 28, 2020
|
|
8SCY
 
 | | Lysozyme crystallized in cyclic olefin copolymer-based microfluidic chips | | Descriptor: | Lysozyme C | | Authors: | Liu, Z, Gu, K, Shelby, M.L, Gilbile, D, Lyubimov, A.Y, Russi, S, Cohen, A.E, Coleman, M.A, Frank, M, Kuhl, T.L. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A user-friendly plug-and-play cyclic olefin copolymer-based microfluidic chip for room-temperature, fixed-target serial crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8SIL
 
 | | Lysozyme crystallized in cyclic olefin copolymer-based microfluidic chips | | Descriptor: | Lysozyme C | | Authors: | Liu, Z, Gu, K, Shelby, M.L, Gilbile, D, Lyubimov, A.Y, Russi, S, Cohen, A.E, Coleman, M.A, Frank, M, Kuhl, T.L, Botha, S, Poitevin, F, Sierra, R.G. | | Deposit date: | 2023-04-16 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A user-friendly plug-and-play cyclic olefin copolymer-based microfluidic chip for room-temperature, fixed-target serial crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
1I44
 
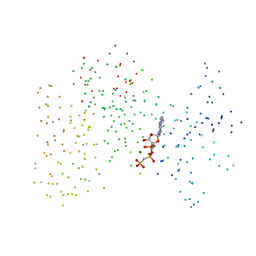 | | CRYSTALLOGRAPHIC STUDIES OF AN ACTIVATION LOOP MUTANT OF THE INSULIN RECEPTOR TYROSINE KINASE | | Descriptor: | INSULIN RECEPTOR, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Till, J.H, Ablooglu, A.J, Frankel, M, Kohanski, R.A, Hubbard, S.R. | | Deposit date: | 2001-02-19 | | Release date: | 2001-03-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic and solution studies of an activation loop mutant of the insulin receptor tyrosine kinase: insights into kinase mechanism.
J.Biol.Chem., 276, 2001
|
|
4OKO
 
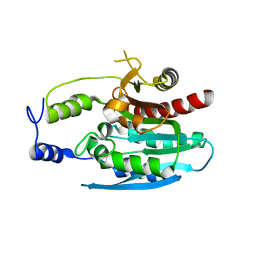 | | Crystal structure of Francisella tularensis REP34 (Rapid Encystment Phenotype Protein 34 KDa) | | Descriptor: | ACETATE ION, Rapid Encystment Phenotype Protein 34 KDa, ZINC ION | | Authors: | Feld, G.K, Segelke, B.W, Corzett, M.H, Hunter, M.S, Frank, M, Rasley, A. | | Deposit date: | 2014-01-22 | | Release date: | 2014-09-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | Structure and Function of REP34 Implicates Carboxypeptidase Activity in Francisella tularensis Host Cell Invasion.
J.Biol.Chem., 289, 2014
|
|
3TRK
 
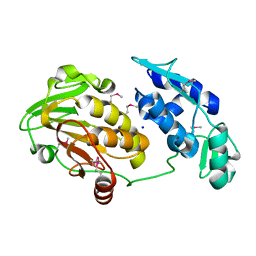 | | Structure of the Chikungunya virus nsP2 protease | | Descriptor: | Nonstructural polyprotein, SODIUM ION | | Authors: | Cheung, J, Franklin, M, Mancia, F, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Structure of the Chikungunya virus nsP2 protease
To be Published
|
|
4GZK
 
 | |
3P3K
 
 | | The crystal structure of translationally controlled tumor protein (TCTP) of Plasmodium falciparum | | Descriptor: | Translationally-controlled tumor protein homolog | | Authors: | Eichhorn, T, Winter, D, Dirdjaja, N, Frank, M, Krauth-Siegel, L, Granzin, J, Efferth, T. | | Deposit date: | 2010-10-05 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Molecular interaction of artemisinin with translationally controlled tumor protein (TCTP) of Plasmodium falciparum.
Biochem Pharmacol, 85, 2013
|
|
7K9P
 
 | | Room temperature structure of NSP15 Endoribonuclease from SARS CoV-2 solved using SFX. | | Descriptor: | CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Botha, S, Jernigan, R, Chen, J, Coleman, M.A, Frank, M, Grant, T.D, Hansen, D.T, Ketawala, G, Logeswaran, D, Martin-Garcia, J, Nagaratnam, N, Raj, A.L.L.X, Shelby, M, Yang, J.-H, Yung, M.C, Fromme, P. | | Deposit date: | 2020-09-29 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Room-temperature structural studies of SARS-CoV-2 protein NendoU with an X-ray free-electron laser.
Structure, 2022
|
|
