7OK0
 
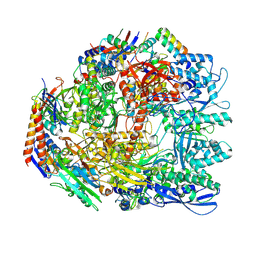 | | Cryo-EM structure of the Sulfolobus acidocaldarius RNA polymerase at 2.88 A | | Descriptor: | Conserved protein, DNA-directed RNA polymerase subunit A', DNA-directed RNA polymerase subunit A'', ... | | Authors: | Pilotto, S, Fouqueau, T, Lukoyanova, N, Sheppard, C, Lucas-Staat, S, Diaz-Santin, L.M, Matelska, D, Prangishvili, D, Cheung, A.C.M, Werner, F. | | Deposit date: | 2021-05-17 | | Release date: | 2021-08-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of RNA polymerase inhibition by viral and host factors.
Nat Commun, 12, 2021
|
|
7OQY
 
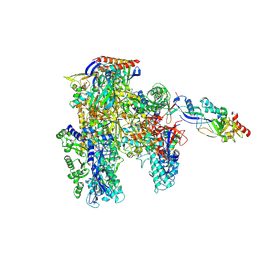 | | Cryo-EM structure of the cellular negative regulator TFS4 bound to the archaeal RNA polymerase | | Descriptor: | Conserved protein, DNA-directed RNA polymerase subunit A', DNA-directed RNA polymerase subunit A'', ... | | Authors: | Pilotto, S, Fouqueau, T, Lukoyanova, N, Sheppard, C, Lucas-Staat, S, Diaz-Santin, L.M, Matelska, D, Prangishvili, D, Cheung, A.C.M, Werner, F. | | Deposit date: | 2021-06-04 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structural basis of RNA polymerase inhibition by viral and host factors.
Nat Commun, 12, 2021
|
|
7OQ4
 
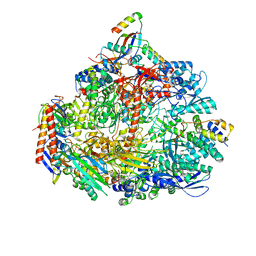 | | Cryo-EM structure of the ATV RNAP Inhibitory Protein (RIP) bound to the DNA-binding channel of the host's RNA polymerase | | Descriptor: | Conserved protein, DNA-directed RNA polymerase subunit A', DNA-directed RNA polymerase subunit A'', ... | | Authors: | Pilotto, S, Fouqueau, T, Lukoyanova, N, Sheppard, C, Lucas-Staat, S, Diaz-Santin, L.M, Matelska, D, Prangishvili, D, Cheung, A.C.M, Werner, F. | | Deposit date: | 2021-06-02 | | Release date: | 2021-08-25 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural basis of RNA polymerase inhibition by viral and host factors.
Nat Commun, 12, 2021
|
|
8Q3B
 
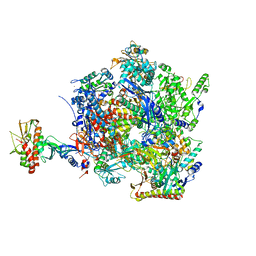 | | The closed state of the ASFV apo-RNA polymerase | | Descriptor: | DNA-directed RNA polymerase RPB1 homolog, DNA-directed RNA polymerase RPB10 homolog, DNA-directed RNA polymerase RPB2 homolog, ... | | Authors: | Pilotto, S, Sykora, M, Cackett, G, Werner, F. | | Deposit date: | 2023-08-03 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structure of the recombinant RNA polymerase from African Swine Fever Virus.
Nat Commun, 15, 2024
|
|
8Q3K
 
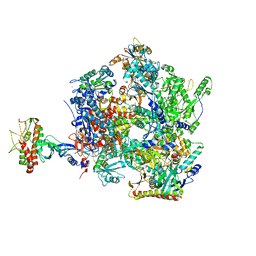 | | The open state of the ASFV apo-RNA polymerase | | Descriptor: | DNA-directed RNA polymerase RPB1 homolog, DNA-directed RNA polymerase RPB10 homolog, DNA-directed RNA polymerase RPB2 homolog, ... | | Authors: | Pilotto, S, Sykora, M, Cackett, G, Werner, F. | | Deposit date: | 2023-08-04 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structure of the recombinant RNA polymerase from African Swine Fever Virus.
Nat Commun, 15, 2024
|
|
5L3D
 
 | | Human LSD1/CoREST: LSD1 Y761H mutation | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, REST corepressor 1 | | Authors: | Pilotto, S, Speranzini, V, Marabelli, C, Mattevi, A. | | Deposit date: | 2016-04-06 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | LSD1/KDM1A mutations associated to a newly described form of intellectual disability impair demethylase activity and binding to transcription factors.
Hum.Mol.Genet., 25, 2016
|
|
5L3B
 
 | | Human LSD1/CoREST: LSD1 D556G mutation | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, REST corepressor 1 | | Authors: | Pilotto, S, Speranzini, V, Marabelli, C. | | Deposit date: | 2016-04-06 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | LSD1/KDM1A mutations associated to a newly described form of intellectual disability impair demethylase activity and binding to transcription factors.
Hum.Mol.Genet., 25, 2016
|
|
5L3C
 
 | | Human LSD1/CoREST: LSD1 E379K mutation | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, REST corepressor 1 | | Authors: | Pilotto, S, Speranzini, V, Marabelli, C, Mattevi, A. | | Deposit date: | 2016-04-06 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | LSD1/KDM1A mutations associated to a newly described form of intellectual disability impair demethylase activity and binding to transcription factors.
Hum.Mol.Genet., 25, 2016
|
|
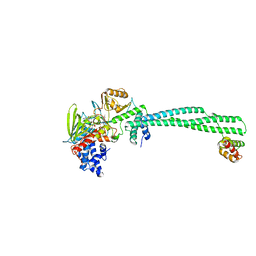
9FJL
 
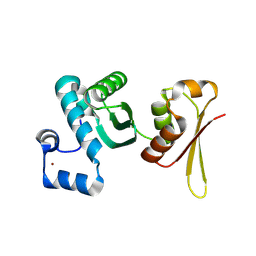 | |
2XAJ
 
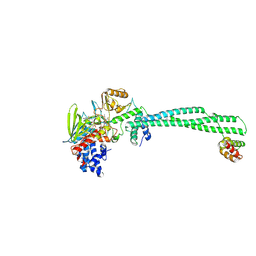 | | Crystal structure of LSD1-CoREST in complex with (-)-trans-2- phenylcyclopropyl-1-amine | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, PHENYLETHYLENECARBOXYLIC ACID, ... | | Authors: | Binda, C, Valente, S, Romanenghi, M, Pilotto, S, Cirilli, R, Karytinos, A, Ciossani, G, Botrugno, O.A, Forneris, F, Tardugno, M, Edmondson, D.E, Minucci, S, Mattevi, A, Mai, A. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Biochemical, Structural, and Biological Evaluation of Tranylcypromine Derivatives as Inhibitors of Histone Demethylases Lsd1 and Lsd2.
J.Am.Chem.Soc., 132, 2010
|
|
4BAY
 
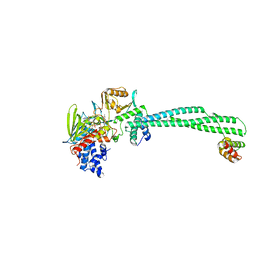 | | Phosphomimetic mutant of LSD1-8a splicing variant in complex with CoREST | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1 | | Authors: | Toffolo, E, Paganini, L, Rusconi, F, Tortorici, M, Pilotto, S, Verpelli, C, Tedeschi, G, Maffioli, E, Sala, C, Mattevi, A, Battaglioli, E. | | Deposit date: | 2012-09-17 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Phosphorylation of Neuronal Lysine-Specific Demethylase 1Lsd1/Kdm1A Impairs Transcriptional Repression by Regulating Interaction with Corest and Histone Deacetylases Hdac1/2.
J.Neurochem., 128, 2014
|
|
6R1T
 
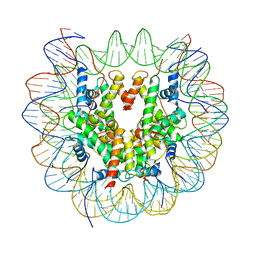 | | Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 1, free nuclesome | | Descriptor: | DNA (147-MER), HISTONE H2A, Histone H2A, ... | | Authors: | Marabelli, C, Pilotto, S, Chittori, S, Subramaniam, S, Mattevi, A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.02 Å) | | Cite: | A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep, 27, 2019
|
|
6R1U
 
 | | Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 2 | | Descriptor: | DNA (147-MER), FLAVIN-ADENINE DINUCLEOTIDE, Histone H2A, ... | | Authors: | Marabelli, C, Pilotto, S, Chittori, S, Subramaniam, S, Mattevi, A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep, 27, 2019
|
|
6R25
 
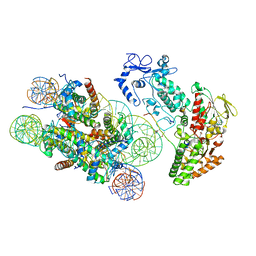 | | Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 3 | | Descriptor: | DNA (147-MER), FLAVIN-ADENINE DINUCLEOTIDE, H2B, ... | | Authors: | Marabelli, C, Pilotto, S, Chittori, S, Subramaniam, S, Mattevi, A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-04-24 | | Method: | ELECTRON MICROSCOPY (4.61 Å) | | Cite: | A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep, 27, 2019
|
|
3ZN1
 
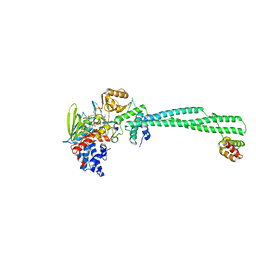 | | LSD1-CoREST in complex with PRLYLV peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, PEPTIDE, ... | | Authors: | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | Deposit date: | 2013-02-13 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
3ZMV
 
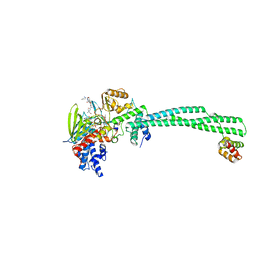 | | LSD1-CoREST in complex with PLSFLV peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, PKSFLV PEPTIDE, ... | | Authors: | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | Deposit date: | 2013-02-12 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
3ZMU
 
 | | LSD1-CoREST in complex with PKSFLV peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, PKSFLV PEPTIDE, ... | | Authors: | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | Deposit date: | 2013-02-12 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
3ZN0
 
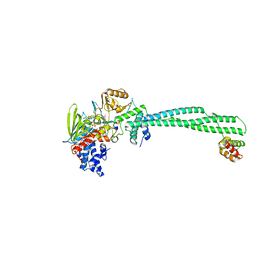 | | LSD1-CoREST in complex with PRSFAA peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, PEPTIDE, ... | | Authors: | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | Deposit date: | 2013-02-13 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
3ZMZ
 
 | | LSD1-CoREST in complex with PRSFAV peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, PEPTIDE, ... | | Authors: | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | Deposit date: | 2013-02-13 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
3ZMS
 
 | | LSD1-CoREST in complex with INSM1 peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, INSULINOMA-ASSOCIATED PROTEIN 1, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, ... | | Authors: | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | Deposit date: | 2013-02-12 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
3ZMT
 
 | | LSD1-CoREST in complex with PRSFLV peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, PEPTIDE, ... | | Authors: | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | Deposit date: | 2013-02-12 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
2XAG
 
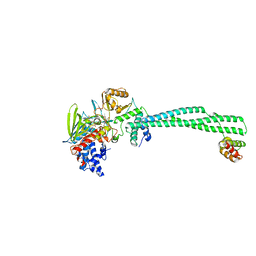 | | Crystal structure of LSD1-CoREST in complex with para-bromo-(-)-trans- 2-phenylcyclopropyl-1-amine | | Descriptor: | 3-(4-BROMOPHENYL)PROPANAMIDE, FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, ... | | Authors: | Binda, C, Valente, S, Romanenghi, M, Pilotto, S, Cirilli, R, Karytinos, A, Ciossani, G, Botrugno, O.A, Forneris, F, Tardugno, M, Edmondson, D.E, Minucci, S, Mattevi, A, Mai, A. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Biochemical, Structural, and Biological Evaluation of Tranylcypromine Derivatives as Inhibitors of Histone Demethylases Lsd1 and Lsd2.
J.Am.Chem.Soc., 132, 2010
|
|
2XAF
 
 | | Crystal structure of LSD1-CoREST in complex with para-bromo-(+)-cis-2- phenylcyclopropyl-1-amine | | Descriptor: | 3-(4-BROMOPHENYL)PROPANAMIDE, FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, ... | | Authors: | Binda, C, Valente, S, Romanenghi, M, Pilotto, S, Cirilli, R, Karytinos, A, Ciossani, G, Botrugno, O.A, Forneris, F, Tardugno, M, Edmondson, D.E, Minucci, S, Mattevi, A, Mai, A. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Biochemical, Structural, and Biological Evaluation of Tranylcypromine Derivatives as Inhibitors of Histone Demethylases Lsd1 and Lsd2.
J.Am.Chem.Soc., 132, 2010
|
|
2XAQ
 
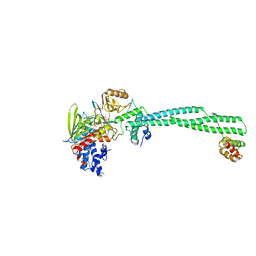 | | Crystal structure of LSD1-CoREST in complex with a tranylcypromine derivative (MC2584, 13b) | | Descriptor: | 3-{4-[(PHENYLCARBONYL)AMINO]PHENYL}PROPANOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, ... | | Authors: | Binda, C, Valente, S, Romanenghi, M, Pilotto, S, Cirilli, R, Karytinos, A, Ciossani, G, Botrugno, O.A, Forneris, F, Tardugno, M, Edmondson, D.E, Minucci, S, Mattevi, A, Mai, A. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Biochemical, Structural, and Biological Evaluation of Tranylcypromine Derivatives as Inhibitors of Histone Demethylases Lsd1 and Lsd2.
J.Am.Chem.Soc., 132, 2010
|
|
2XAH
 
 | | Crystal structure of LSD1-CoREST in complex with (+)-trans-2- phenylcyclopropyl-1-amine | | Descriptor: | 3-PHENYLPROPANAL, FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, ... | | Authors: | Binda, C, Valente, S, Romanenghi, M, Pilotto, S, Cirilli, R, Karytinos, A, Ciossani, G, Botrugno, O.A, Forneris, F, Tardugno, M, Edmondson, D.E, Minucci, S, Mattevi, A, Mai, A. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Biochemical, Structural, and Biological Evaluation of Tranylcypromine Derivatives as Inhibitors of Histone Demethylases Lsd1 and Lsd2.
J.Am.Chem.Soc., 132, 2010
|
|
