1IB1
 
 | | CRYSTAL STRUCTURE OF THE 14-3-3 ZETA:SEROTONIN N-ACETYLTRANSFERASE COMPLEX | | Descriptor: | 14-3-3 ZETA ISOFORM, COA-S-ACETYL TRYPTAMINE, SEROTONIN N-ACETYLTRANSFERASE | | Authors: | Obsil, T, Ghirlando, R, Klein, D.C, Ganguly, S, Dyda, F. | | Deposit date: | 2001-03-26 | | Release date: | 2001-05-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the 14-3-3zeta:serotonin N-acetyltransferase complex. a role for scaffolding in enzyme regulation.
Cell(Cambridge,Mass.), 105, 2001
|
|
8QGY
 
 | |
6EWW
 
 | |
6QVW
 
 | | Solution structure of the free FOXO1 DNA binding domain | | Descriptor: | Forkhead box protein O1 | | Authors: | Psenakova, K, Obsil, T, Veverka, V, Obsilova, V, Kohoutova, K. | | Deposit date: | 2019-03-05 | | Release date: | 2019-09-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Forkhead Domains of FOXO Transcription Factors Differ in both Overall Conformation and Dynamics.
Cells, 8, 2019
|
|
6QK8
 
 | |
7NMZ
 
 | |
6EJL
 
 | |
3L2C
 
 | | Crystal Structure of the DNA Binding Domain of FOXO4 Bound to DNA | | Descriptor: | FOXO consensus binding sequence, minus strand, plus strand, ... | | Authors: | Boura, E, Silhan, J, Sulc, M, Brynda, J, Obsilova, V, Obsil, T. | | Deposit date: | 2009-12-15 | | Release date: | 2009-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.868 Å) | | Cite: | Structure of the human FOXO4-DBD-DNA complex at 1.9 A resolution reveals new details of FOXO binding to the DNA
Acta Crystallogr.,Sect.D, 66, 2010
|
|
5JTA
 
 | |
6S9K
 
 | | Structure of 14-3-3 gamma in complex with caspase-2 peptide containing 14-3-3 binding motif Ser139 and NLS | | Descriptor: | 1,1,1,3,3,3-hexafluoropropan-2-ol, 14-3-3 protein gamma, Caspase-2 | | Authors: | Alblova, M, Obsil, T, Obsilova, V. | | Deposit date: | 2019-07-15 | | Release date: | 2020-01-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 14-3-3 protein binding blocks the dimerization interface of caspase-2.
Febs J., 287, 2020
|
|
6SAD
 
 | |
5NIS
 
 | |
5N6N
 
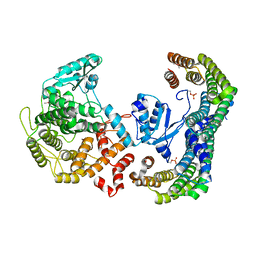 | | CRYSTAL STRUCTURE OF THE 14-3-3:NEUTRAL TREHALASE NTH1 COMPLEX | | Descriptor: | CALCIUM ION, Neutral trehalase, Protein BMH1, ... | | Authors: | Alblova, M, Smidova, A, Obsilova, V, Obsil, T. | | Deposit date: | 2017-02-15 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Molecular basis of the 14-3-3 protein-dependent activation of yeast neutral trehalase Nth1.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5M4A
 
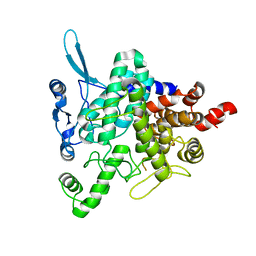 | | Neutral trehalase Nth1 from Saccharomyces cerevisiae in complex with trehalose | | Descriptor: | Neutral trehalase, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Smidova, A, Alblova, M, Obsilova, V, Obsil, T. | | Deposit date: | 2016-10-18 | | Release date: | 2017-11-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular basis of the 14-3-3 protein-dependent activation of yeast neutral trehalase Nth1.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2OJ4
 
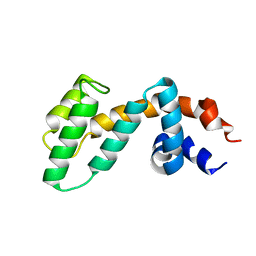 | | Crystal structure of RGS3 RGS domain | | Descriptor: | Regulator of G-protein signaling 3 | | Authors: | Boura, E, Obsil, T. | | Deposit date: | 2007-01-12 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 14-3-3 protein interacts with and affects the structure of RGS domain of regulator of G protein signaling 3 (RGS3).
J.Struct.Biol., 170, 2010
|
|
6Y4K
 
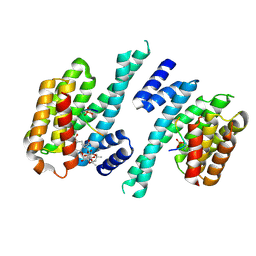 | |
6Y6B
 
 | | Crystal structure of human 14-3-3 gamma in complex with CaMKK2 14-3-3 binding motif Ser100 and 16-OMe-Fusicoccin H | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[[(1~{S},3~{R},6~{S},7~{S},8~{R},9~{R},10~{R},11~{R},14~{S})-14-(me thoxymethyl)-3,10-dimethyl-9-oxidanyl-6-propan-2-yl-8-tricyclo[9.3.0.0^{3,7}]tetradecanyl]oxy]oxane-3,4,5-triol, 14-3-3 protein gamma, Calcium/calmodulin-dependent protein kinase kinase 2 | | Authors: | Lentini Santo, D, Obsilova, V, Obsil, T. | | Deposit date: | 2020-02-26 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Stabilization of Protein-Protein Interactions between CaMKK2 and 14-3-3 by Fusicoccins.
Acs Chem.Biol., 15, 2020
|
|
6FEL
 
 | |
6GKF
 
 | |
6GKG
 
 | |
6ZBT
 
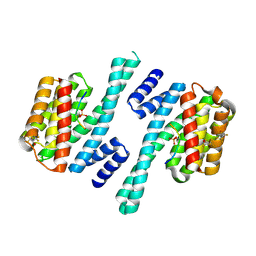 | | Structure of 14-3-3 gamma in complex with Nedd4-2 14-3-3 binding motif Ser342 | | Descriptor: | 1,1,1,3,3,3-hexafluoropropan-2-ol, 14-3-3 protein gamma, E3 ubiquitin-protein ligase NEDD4-like | | Authors: | Joshi, R, Kalabova, D, Obsil, T, Obsilova, V. | | Deposit date: | 2020-06-09 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79948521 Å) | | Cite: | 14-3-3-protein regulates Nedd4-2 by modulating interactions between HECT and WW domains.
Commun Biol, 4, 2021
|
|
6ZC9
 
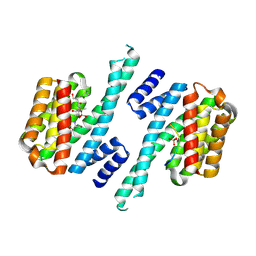 | | Structure of 14-3-3 gamma in complex with Nedd4-2 14-3-3 binding motif Ser448 | | Descriptor: | 1,1,1,3,3,3-hexafluoropropan-2-ol, 14-3-3 protein gamma, E3 ubiquitin-protein ligase NEDD4-like | | Authors: | Pohl, P, Kalabova, D, Obsil, T, Obsilova, V. | | Deposit date: | 2020-06-10 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.89895988 Å) | | Cite: | 14-3-3-protein regulates Nedd4-2 by modulating interactions between HECT and WW domains.
Commun Biol, 4, 2021
|
|
7A6Y
 
 | |
7A6R
 
 | |
6QIU
 
 | | Crystal structure of 14-3-3 sigma in complex with Ataxin-1 Ser776 phosphopeptide | | Descriptor: | 14-3-3 protein sigma, Ataxin-1 phosphopeptide, CHLORIDE ION, ... | | Authors: | Leysen, S, Milroy, L.G, Davis, J.M, Brunsveld, L, Ottmann, C. | | Deposit date: | 2019-01-21 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structural insights into the cytoplasmic chaperone effect of 14-3-3 proteins on Ataxin-1
To Be Published
|
|
