6R6H
 
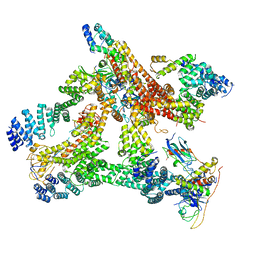 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Morris, E.P, Faull, S.V, Lau, A.M.C, Politis, A, Beuron, F, Cronin, N. | | Deposit date: | 2019-03-27 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
6R7F
 
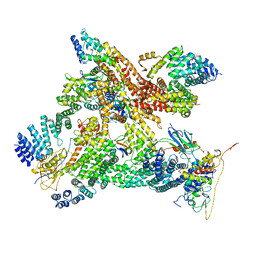 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Faull, S.V, Lau, A.M.C, Martens, C, Ahdash, Z, Yebenes, H, Schmidt, C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | Deposit date: | 2019-03-28 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
4UX8
 
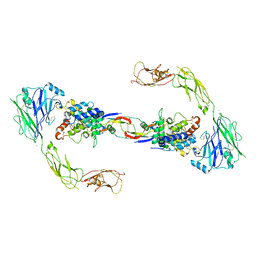 | | RET recognition of GDNF-GFRalpha1 ligand by a composite binding site promotes membrane-proximal self-association | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GDNF FAMILY RECEPTOR ALPHA-1, ... | | Authors: | Goodman, K, Kjaer, S, Beuron, F, Knowles, P, Nawrotek, A, Burns, E, Purkiss, A, George, R, Santoro, M, Morris, E.P, McDonald, N.Q. | | Deposit date: | 2014-08-19 | | Release date: | 2014-10-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (24 Å) | | Cite: | Ret Recognition of Gdnf-Gfralpha1 Ligand by a Composite Binding Site Promotes Membrane-Proximal Self-Association.
Cell Rep., 8, 2014
|
|
5A0Q
 
 | |
8ALY
 
 | | Cryo-EM structure of human tankyrase 2 SAM-PARP filament (G1032W mutant) | | Descriptor: | Poly [ADP-ribose] polymerase tankyrase-2, ZINC ION | | Authors: | Mariotti, L, Inian, O, Desfosses, A, Beuron, F, Morris, E.P, Guettler, S. | | Deposit date: | 2022-08-01 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis of tankyrase activation by polymerization.
Nature, 612, 2022
|
|
5MVA
 
 | |
5MVY
 
 | | Thin Filament at low calcium concentration | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle | | Authors: | Paul, D.M, Squire, J.M, Morris, E.P. | | Deposit date: | 2017-01-17 | | Release date: | 2018-02-14 | | Last modified: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (28.4 Å) | | Cite: | Relaxed and active thin filament structures; a new structural basis for the regulatory mechanism.
J. Struct. Biol., 197, 2017
|
|
4ADY
 
 | | Crystal structure of 26S proteasome subunit Rpn2 | | Descriptor: | 26S PROTEASOME REGULATORY SUBUNIT RPN2 | | Authors: | Kulkarni, K, He, J, Da Fonseca, P.C.A, Krutauz, D, Glickman, M.H, Barford, D, Morris, E.P. | | Deposit date: | 2012-01-04 | | Release date: | 2012-03-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structure of the 26S Proteasome Subunit Rpn2 Reveals its Pc Repeat Domain as a Closed Toroid of Two Concentric Alpha-Helical Rings
Structure, 20, 2012
|
|
6R7N
 
 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Faull, S.V, Lau, A.M.C, Martens, C, Ahdash, Z, Yebenes, H, Schmidt, C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | Deposit date: | 2019-03-29 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
6R7H
 
 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Faull, S.V, Lau, A.M.C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | Deposit date: | 2019-03-28 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
6R7I
 
 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Faull, S.F, Lau, A.M.C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | Deposit date: | 2019-03-28 | | Release date: | 2019-08-28 | | Last modified: | 2019-09-04 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
6SO3
 
 | | The interacting head motif in insect flight muscle myosin thick filaments | | Descriptor: | Myosin 2 essential light chain striated muscle, Myosin 2 heavy chain striated muscle, Myosin 2 regulatory light chain striated muscle | | Authors: | Morris, E.P, Knupp, C, Squire, J.M. | | Deposit date: | 2019-08-28 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | The Interacting Head Motif Structure Does Not Explain the X-Ray Diffraction Patterns in Relaxed Vertebrate (Bony Fish) Skeletal Muscle and Insect (Lethocerus) Flight Muscle.
Biology (Basel), 8, 2019
|
|
6SXB
 
 | | XPF-ERCC1 Cryo-EM Structure, DNA-Bound form | | Descriptor: | DNA (5'-D(*TP*CP*AP*GP*CP*AP*TP*CP*TP*G)-3'), DNA (5'-D(P*CP*AP*GP*AP*TP*GP*CP*TP*GP*A)-3'), DNA excision repair protein ERCC-1, ... | | Authors: | Jones, M.L, Briggs, D.C, McDonald, N.Q. | | Deposit date: | 2019-09-25 | | Release date: | 2020-03-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Cryo-EM structures of the XPF-ERCC1 endonuclease reveal how DNA-junction engagement disrupts an auto-inhibited conformation.
Nat Commun, 11, 2020
|
|
6SXA
 
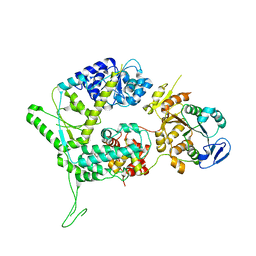 | | XPF-ERCC1 Cryo-EM Structure, Apo-form | | Descriptor: | DNA excision repair protein ERCC-1, DNA repair endonuclease XPF | | Authors: | Jones, M.L, Briggs, D.C, McDonald, N.Q. | | Deposit date: | 2019-09-25 | | Release date: | 2020-03-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of the XPF-ERCC1 endonuclease reveal how DNA-junction engagement disrupts an auto-inhibited conformation.
Nat Commun, 11, 2020
|
|
5TBY
 
 | | HUMAN BETA CARDIAC HEAVY MEROMYOSIN INTERACTING-HEADS MOTIF OBTAINED BY HOMOLOGY MODELING (USING SWISS-MODEL) OF HUMAN SEQUENCE FROM APHONOPELMA HOMOLOGY MODEL (PDB-3JBH), RIGIDLY FITTED TO HUMAN BETA-CARDIAC NEGATIVELY STAINED THICK FILAMENT 3D-RECONSTRUCTION (EMD-2240) | | Descriptor: | Myosin light chain 3, Myosin regulatory light chain 2, ventricular/cardiac muscle isoform, ... | | Authors: | ALAMO, L, WARE, J.S, PINTO, A, GILLILAN, R.E, SEIDMAN, J.G, SEIDMAN, C.E, PADRON, R. | | Deposit date: | 2016-09-13 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Effects of myosin variants on interacting-heads motif explain distinct hypertrophic and dilated cardiomyopathy phenotypes.
Elife, 6, 2017
|
|
