1CGU
 
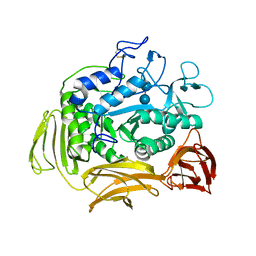 | | CATALYTIC CENTER OF CYCLODEXTRIN GLYCOSYLTRANSFERASE DERIVED FROM X-RAY STRUCTURE ANALYSIS COMBINED WITH SITE-DIRECTED MUTAGENESIS | | Descriptor: | CALCIUM ION, CYCLODEXTRIN GLYCOSYL-TRANSFERASE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Klein, C, Hollender, J, Bender, H, Schulz, G.E. | | Deposit date: | 1992-06-10 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Catalytic center of cyclodextrin glycosyltransferase derived from X-ray structure analysis combined with site-directed mutagenesis.
Biochemistry, 31, 1992
|
|
1CGT
 
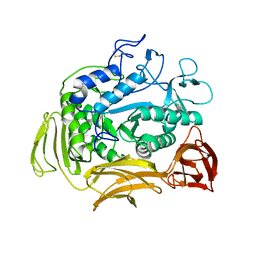 | |
5M5E
 
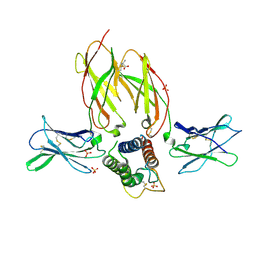 | | Crystal structure of a interleukin-2 variant in complex with interleukin-2 receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Klein, C, Freimoser-Grundschober, A, Waldhauer, I, Stihle, M, Birk, M, Benz, J. | | Deposit date: | 2016-10-21 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cergutuzumab amunaleukin (CEA-IL2v), a CEA-targeted IL-2 variant-based immunocytokine for combination cancer immunotherapy: Overcoming limitations of aldesleukin and conventional IL-2-based immunocytokines.
Oncoimmunology, 6, 2017
|
|
4V7F
 
 | | Arx1 pre-60S particle. | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Leidig, C, Thoms, M, Holdermann, I, Bradatsch, B, Berninghausen, O, Bange, G, Sinning, I, Hurt, E, Beckmann, R. | | Deposit date: | 2013-12-10 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | 60S ribosome biogenesis requires rotation of the 5S ribonucleoprotein particle.
Nat Commun, 5, 2014
|
|
1GAR
 
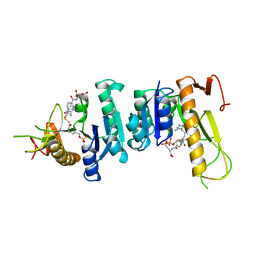 | | TOWARDS STRUCTURE-BASED DRUG DESIGN: CRYSTAL STRUCTURE OF A MULTISUBSTRATE ADDUCT COMPLEX OF GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE AT 1.96 ANGSTROMS RESOLUTION | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE, N-[4-[[3-(2,4-DIAMINO-1,6-DIHYDRO-6-OXO-4-PYRIMIDINYL)-PROPYL]-[2-((2-OXO-2-((4-PHOSPHORIBOXY)-BUTYL)-AMINO)-ETHYL)-THIO-ACETYL]-AMINO]BENZOYL]-1-GLUTAMIC ACID | | Authors: | Wilson, I.A, Klein, C, Chen, P, Arevalo, J.H. | | Deposit date: | 1994-12-08 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Towards structure-based drug design: crystal structure of a multisubstrate adduct complex of glycinamide ribonucleotide transformylase at 1.96 A resolution.
J.Mol.Biol., 249, 1995
|
|
8BFB
 
 | | Sarkosyl-extracted AppNL-G-F Abeta42 fibril structure (Methoxy-X04-labelled mice) | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Wilkinson, M, Leistner, C, Burgess, A, Goodfellow, S, Deuchars, S, Ranson, N.A, Radford, S.E, Frank, R.A.W. | | Deposit date: | 2022-10-24 | | Release date: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The in-tissue molecular architecture of beta-amyloid pathology in the mammalian brain.
Nat Commun, 14, 2023
|
|
8BFA
 
 | | Sarkosyl-extracted AppNL-G-F Abeta42 fibril structure | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Wilkinson, M, Leistner, C, Burgess, A, Goodfellow, S, Deuchars, S, Ranson, N.A, Radford, S.E, Frank, R.A.W. | | Deposit date: | 2022-10-24 | | Release date: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The in-tissue molecular architecture of beta-amyloid pathology in the mammalian brain.
Nat Commun, 14, 2023
|
|
3TJT
 
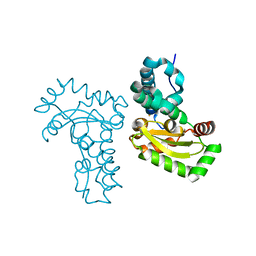 | | Crystal Structure Analysis of the superoxide dismutase from Clostridium difficile | | Descriptor: | FE (III) ION, Superoxide dismutase | | Authors: | Li, W, Lei, C, Ying, T.L, Wang, H.F, Tan, X.S. | | Deposit date: | 2011-08-25 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal Structure of the superoxide dismutase from Clostridium difficile
To be Published
|
|
2VN6
 
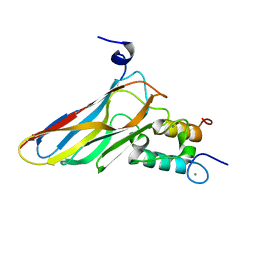 | | The Clostridium cellulolyticum dockerin displays a dual binding mode for its cohesin partner | | Descriptor: | CALCIUM ION, ENDOGLUCANASE A, SCAFFOLDING PROTEIN | | Authors: | Pinheiro, B.A, Prates, J.A.M, Proctor, M.R, Gilbert, H.J, Davies, G.J, Money, V.A, Martinez-Fleites, C, Bayer, E.A, Fontes, C.M.G.A, Fierobe, H.P. | | Deposit date: | 2008-01-31 | | Release date: | 2008-05-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | The Clostridium Cellulolyticum Dockerin Displays a Dual Binding Mode for its Cohesin Partner.
J.Biol.Chem., 283, 2008
|
|
4V0G
 
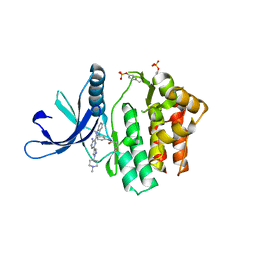 | | JAK3 in complex with a covalent EGFR inhibitor | | Descriptor: | N-[3-(2-{3-amino-6-[1-(1-methylpiperidin-4-yl)-1H-pyrazol-4-yl]pyrazin-2-yl}-1H-benzimidazol-1-yl)phenyl]propanamide, TYROSINE-PROTEIN KINASE JAK3 | | Authors: | Debreczeni, J.E, Hennessy, E.J, Chuaquini, C, Ashton, S, Coclough, N, Cross, D.A.E, Eberlein, C, Gingipalli, L, Klinowska, T.C.M, Orme, J.P, Sha, L, Wu, X. | | Deposit date: | 2014-09-16 | | Release date: | 2016-01-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Utilisation of Structure Based Design to Identify Novel, Irreversible Inhibitors of the Epidermal Growth Factor Receptor (Egfr) Harboring the Gatekeeper T790M Mutation
To be Published
|
|
4XYC
 
 | | NANOMOLAR INHIBITORS OF MYCOBACTERIUM TUBERCULOSIS GLUTAMINE SYNTHETASE 1: SYNTHESIS, BIOLOGICAL EVALUATION AND X-RAY CRYSTALLOGRAPHIC STUDIES | | Descriptor: | 9-phenyl-4H-imidazo[1,2-a]indeno[1,2-e]pyrazin-4-one, Glutamine synthetase 1 | | Authors: | Couturier, C, Silve, S, Morales, R, Ppessegue, B, Llopart, S, Nair, A, Bauer, A, Scheiper, B, poeverlein, c, Ganzhorn, A, Lagrange, S, Bacque, E. | | Deposit date: | 2015-02-02 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Nanomolar inhibitors of Mycobacterium tuberculosis glutamine synthetase 1: Synthesis, biological evaluation and X-ray crystallographic studies.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
1TT0
 
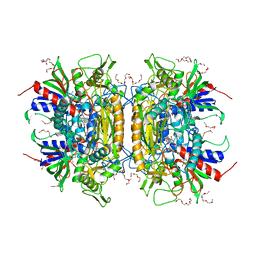 | | Crystal Structure of Pyranose 2-Oxidase | | Descriptor: | ACETATE ION, DODECAETHYLENE GLYCOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Hallberg, B.M, Leitner, C, Haltrich, D, Divne, C. | | Deposit date: | 2004-06-21 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the 270 kDa homotetrameric lignin-degrading enzyme pyranose 2-oxidase
J.Mol.Biol., 341, 2004
|
|
4LI5
 
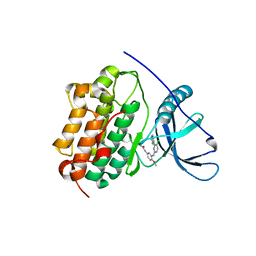 | | EGFR-K IN COMPLEX WITH N-[3-[[5-chloro-4-(1H-indol-3-yl)pyrimidin-2-yl]amino]-4-methoxy-phenyl] Prop-2-enamide | | Descriptor: | Epidermal growth factor receptor, N-(3-{[5-chloro-4-(1H-indol-3-yl)pyrimidin-2-yl]amino}-4-methoxyphenyl)propanamide, SODIUM ION | | Authors: | Debreczeni, J.E, Seiffert, G.B, Kiefersauer, R, Augustin, M, Nagel, S, Ward, R, Anderton, M, Ashton, S, Bethel, P, Box, M, Butterworth, S, Colclough, N, Chroley, C, Chuaqui, C, Cross, D, Eberlein, C, Finlay, R, Hill, G, Grist, M, Klinowska, T, Lane, C, Martin, S, Orme, J, Smith, P, Wang, F, Waring, M. | | Deposit date: | 2013-07-02 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure- and Reactivity-Based Development of Covalent Inhibitors of the Activating and Gatekeeper Mutant Forms of the Epidermal Growth Factor Receptor (EGFR).
J.Med.Chem., 56, 2013
|
|
1W18
 
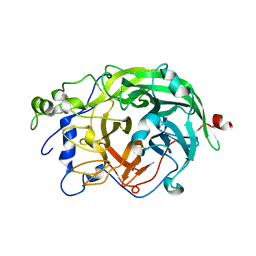 | | Crystal Structure of levansucrase from Gluconacetobacter diazotrophicus | | Descriptor: | LEVANSUCRASE, SULFATE ION | | Authors: | Martinez-Fleites, C, Ortiz-Lombardia, M, Pons, T, Tarbouriech, N, Taylor, E.J, Hernandez, L, Davies, G.J. | | Deposit date: | 2004-06-16 | | Release date: | 2005-05-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Levansucrase from the Gram- Negative Bacterium Gluconacetobacter Diazotrophicus.
Biochem.J., 390, 2005
|
|
1UYW
 
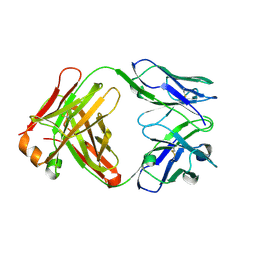 | | Crystal Structure of the antiflavivirus Fab4g2 | | Descriptor: | FAB ANTIBODY HEAVY CHAIN, FAB ANTIBODY LIGHT CHAIN | | Authors: | Martinez-Fleites, C, Ortiz-Lombardia, M, Taylor, E.J, Gil-Valdes, J, Chinea, G, Davies, G. | | Deposit date: | 2004-03-03 | | Release date: | 2005-03-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Antiflavivirus Fab4G2
To be Published
|
|
6FIB
 
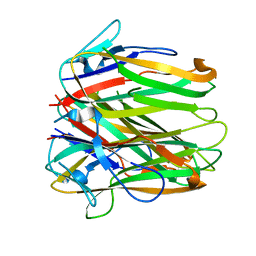 | | Structure of human 4-1BB ligand | | Descriptor: | Tumor necrosis factor ligand superfamily member 9, Tumor necrosis factor ligand superfamily member 9,4-1BBL -CH/CL fusion, Tumor necrosis factor ligand superfamily member 9,Uncharacterized protein | | Authors: | Joseph, C, Claus, C, Ferrara, C, von Hirschheydt, T, Prince, C, Funk, D, Klein, C, Benz, J. | | Deposit date: | 2018-01-17 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Tumor-targeted 4-1BB agonists for combination with T cell bispecific antibodies as off-the-shelf therapy.
Sci Transl Med, 11, 2019
|
|
2VN5
 
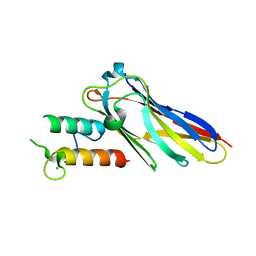 | | The Clostridium cellulolyticum dockerin displays a dual binding mode for its cohesin partner | | Descriptor: | CALCIUM ION, ENDOGLUCANASE A, SCAFFOLDING PROTEIN | | Authors: | Pinheiro, B.A, Prates, J.A.M, Proctor, M.R, Gilbert, H.J, Davies, G.J, Money, V.A, Martinez-Fleites, C, Bayer, E.A, Fontes, C.M.G.A, Fierobe, H.P. | | Deposit date: | 2008-01-31 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Clostridium Cellulolyticum Dockerin Displays a Dual Binding Mode for its Cohesin Partner.
J.Biol.Chem., 283, 2008
|
|
4ZJ0
 
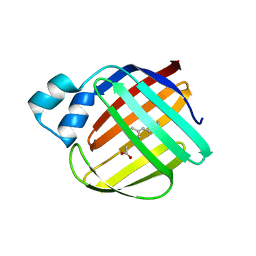 | | The crystal structure of monomer Q108K:K40L:Y60W CRBPII bound to all-trans-retinal | | Descriptor: | ACETATE ION, RETINAL, Retinol-binding protein 2 | | Authors: | Nossoni, Z, Assar, Z, Wang, W, Vasileiou, C, Borhan, B, Geiger, J.H. | | Deposit date: | 2015-04-28 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Domain-Swapped Dimers of Intracellular Lipid-Binding Proteins: Evidence for Ordered Folding Intermediates.
Structure, 24, 2016
|
|
2CN2
 
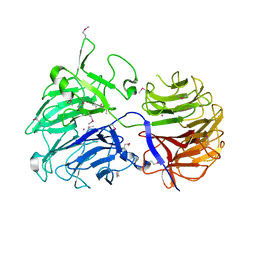 | | Crystal Structures of Clostridium thermocellum Xyloglucanase | | Descriptor: | BETA-1,4-XYLOGLUCAN HYDROLASE, CADMIUM ION | | Authors: | Martinez-Fleites, C, Taylor, E.J, Guerreiro, C.I, Prates, J.A.M, Ferreira, L.M.A, Fontes, C.M.G.A, Baumann, M.J, Brumer, H, Davies, G.J. | | Deposit date: | 2006-05-17 | | Release date: | 2006-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Clostridium Thermocellum Xyloglucanase, Xgh74A, Reveal the Structural Basis for Xyloglucan Recognition and Degradation.
J.Biol.Chem., 281, 2006
|
|
2BO7
 
 | | DISSECTION OF MANNOSYLGLYCERATE SYNTHASE: AN ARCHETYPAL MANNOSYLTRANSFERASE | | Descriptor: | COBALT (II) ION, GUANOSINE-5'-DIPHOSPHATE, MANNOSYLGLYCERATE SYNTHASE | | Authors: | Flint, J, Taylor, E, Yang, M, Bolam, D.N, Tailford, L.E, Martinez-Fleites, C, Dodson, E.J, Davis, B.G, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2005-04-08 | | Release date: | 2005-06-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural dissection and high-throughput screening of mannosylglycerate synthase.
Nat. Struct. Mol. Biol., 12, 2005
|
|
2CN3
 
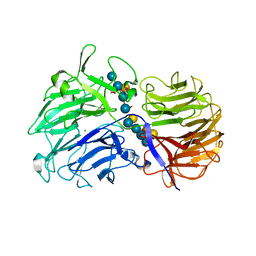 | | Crystal Structures of Clostridium thermocellum Xyloglucanase | | Descriptor: | BETA-1,4-XYLOGLUCAN HYDROLASE, CALCIUM ION, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[beta-D-galactopyranose-(1-2)-alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Martinez-Fleites, C, Taylor, E.J, Guerreiro, C.I.P.D, Prates, J.A.M, Ferreira, L.M.A, Fontes, C.M.G.A, Baumann, M.J, Brumer, H, Davies, G.J. | | Deposit date: | 2006-05-17 | | Release date: | 2006-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structures of Clostridium Thermocellum Xyloglucanase, Xgh74A, Reveal the Structural Basis for Xyloglucan Recognition and Degradation
J.Biol.Chem., 281, 2006
|
|
2Y4M
 
 | | MANNOSYLGLYCERATE SYNTHASE IN COMPLEX WITH GDP-Mannose | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GUANOSINE 5'-(TRIHYDROGEN DIPHOSPHATE), ... | | Authors: | Nielsen, M.M, Suits, M.D.L, Yang, M, Barry, C.S, Martinez-Fleites, C, Tailford, L.E, Flint, J.E, Davis, B.G, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2011-01-07 | | Release date: | 2011-02-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Substrate and Metal Ion Promiscuity in Mannosylglycerate Synthase.
J.Biol.Chem., 286, 2011
|
|
2Y4K
 
 | | MANNOSYLGLYCERATE SYNTHASE IN COMPLEX WITH MG-GDP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Nielsen, M.M, Suits, M.D.L, Yang, M, Barry, C.S, Martinez-Fleites, C, Tailford, L.E, Flint, J.E, Davis, B.G, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2011-01-07 | | Release date: | 2011-02-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Substrate and Metal Ion Promiscuity in Mannosylglycerate Synthase.
J.Biol.Chem., 286, 2011
|
|
2C1X
 
 | | Structure and activity of a flavonoid 3-O glucosyltransferase reveals the basis for plant natural product modification | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, UDP-GLUCOSE FLAVONOID 3-O GLYCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Offen, W, Martinez-Fleites, C, Kiat-Lim, E, Yang, M, Davis, B.G, Tarling, C.A, Ford, C.M, Bowles, D.J, Davies, G.J. | | Deposit date: | 2005-09-22 | | Release date: | 2006-01-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Flavonoid Glucosyltransferase Reveals the Basis for Plant Natural Product Modification.
Embo J., 25, 2006
|
|
2C9Z
 
 | | Structure and activity of a flavonoid 3-0 glucosyltransferase reveals the basis for plant natural product modification | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, UDP GLUCOSE:FLAVONOID 3-O-GLUCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Offen, W, Martinez-Fleites, C, Kiat-Lim, E, Yang, M, Davis, B.G, Tarling, C.A, Ford, C.M, Bowles, D.J, Davies, G.J. | | Deposit date: | 2005-12-15 | | Release date: | 2006-01-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a Flavonoid Glucosyltransferase Reveals the Basis for Plant Natural Product Modification.
Embo J., 25, 2006
|
|
