4HON
 
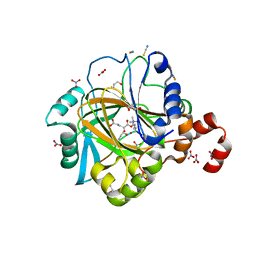 | |
4HOO
 
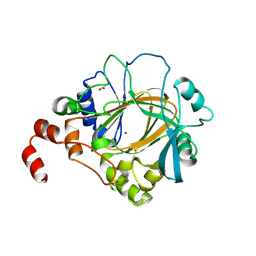 | | Crystal structure of human JMJD2D/KDM4D apoenzyme | | Descriptor: | ACETATE ION, Lysine-specific demethylase 4D, NICKEL (II) ION, ... | | Authors: | Krishnan, S, Trievel, R.C. | | Deposit date: | 2012-10-22 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Structural and Functional Analysis of JMJD2D Reveals Molecular Basis for Site-Specific Demethylation among JMJD2 Demethylases.
Structure, 21, 2013
|
|
8U5Y
 
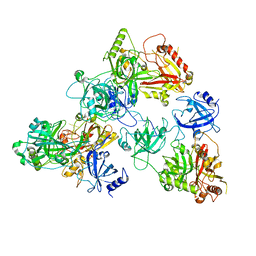 | | human RADX trimer bound to ssDNA | | Descriptor: | DNA (25-MER), RPA-related protein RADX | | Authors: | Balakrishnan, S, Chazin, W.J. | | Deposit date: | 2023-09-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structure of RADX and mechanism for regulation of RAD51 nucleofilaments.
Biorxiv, 2023
|
|
8U61
 
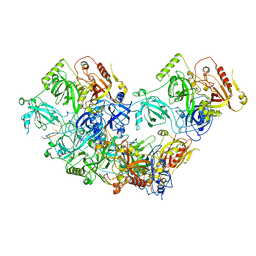 | | Human RADX tetramer bound to ssDNA | | Descriptor: | RPA-related protein RADX, dT25 DNA (25-MER) | | Authors: | Balakrishnan, S, Chazin, W.J. | | Deposit date: | 2023-09-13 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of RADX and mechanism for regulation of RAD51 nucleofilaments.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5XE4
 
 | |
5XEE
 
 | |
4D9T
 
 | | Rsk2 C-terminal Kinase Domain with inhibitor (E)-methyl 3-(4-amino-7-(3-hydroxypropyl)-5-p-tolyl-7H-pyrrolo[2,3-d]pyrimidin-6-yl)-2-cyanoacrylate | | Descriptor: | Ribosomal protein S6 kinase alpha-3, SODIUM ION, methyl (2S)-3-{4-amino-7-[(1E)-3-hydroxyprop-1-en-1-yl]-5-(4-methylphenyl)-7H-pyrrolo[2,3-d]pyrimidin-6-yl}-2-cyanopropanoate | | Authors: | Serafimova, I.M, Pufall, M.A, Krishnan, S, Duda, K, Cohen, M.S, Maglathlin, R.L, McFarland, J.M, Miller, R.M, Frodin, M, Taunton, J. | | Deposit date: | 2012-01-12 | | Release date: | 2012-04-25 | | Last modified: | 2012-05-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Reversible targeting of noncatalytic cysteines with chemically tuned electrophiles.
Nat.Chem.Biol., 8, 2012
|
|
4D9U
 
 | | Rsk2 C-terminal Kinase Domain, (E)-tert-butyl 3-(4-amino-7-(3-hydroxypropyl)-5-p-tolyl-7H-pyrrolo[2,3-d]pyrimidin-6-yl)-2-cyanoacrylate | | Descriptor: | Ribosomal protein S6 kinase alpha-3, SODIUM ION, tert-butyl (2S)-3-[4-amino-7-(3-hydroxypropyl)-5-(4-methylphenyl)-7H-pyrrolo[2,3-d]pyrimidin-6-yl]-2-cyanopropanoate | | Authors: | Serafimova, I.M, Pufall, M.A, Krishnan, S, Duda, K, Cohen, M.S, Maglathlin, R.L, McFarland, J.M, Miller, R.M, Frodin, M, Taunton, J. | | Deposit date: | 2012-01-12 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Reversible targeting of noncatalytic cysteines with chemically tuned electrophiles.
Nat.Chem.Biol., 8, 2012
|
|
4JG6
 
 | | RSK2 CTD bound to 2-cyano-3-(1H-indazol-5-yl)acrylamide | | Descriptor: | (2S)-2-cyano-3-(1H-indazol-5-yl)propanamide, Ribosomal protein S6 kinase alpha-3, SODIUM ION | | Authors: | Miller, R.M, Paavilainen, V.O, Krishnan, S, Serafimova, I.M, Taunton, J. | | Deposit date: | 2013-02-28 | | Release date: | 2013-04-10 | | Last modified: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Electrophilic fragment-based design of reversible covalent kinase inhibitors.
J.Am.Chem.Soc., 135, 2013
|
|
4JG7
 
 | | Structure of RSK2 CTD bound to 3-(3-(1H-pyrrolo[2,3-b]pyridine-3-carbonyl)phenyl)-2-cyanoacrylamide | | Descriptor: | (2R)-2-cyano-3-[3-(1H-pyrrolo[2,3-b]pyridin-3-ylcarbonyl)phenyl]propanamide, Ribosomal protein S6 kinase alpha-3, SODIUM ION | | Authors: | Miller, R.M, Paavilainen, V.O, Krishnan, S, Serafimova, I.M, Taunton, J. | | Deposit date: | 2013-02-28 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.0002 Å) | | Cite: | Electrophilic fragment-based design of reversible covalent kinase inhibitors.
J.Am.Chem.Soc., 135, 2013
|
|
4JG8
 
 | | Structure of RSK2 T493M CTD mutant bound to 2-cyano-N-(1-hydroxy-2-methylpropan-2-yl)-3-(3-(3,4,5-trimethoxyphenyl)-1H-indazol-5-yl)acrylamide | | Descriptor: | (2S)-2-cyano-N-(1-hydroxy-2-methylpropan-2-yl)-3-[3-(3,4,5-trimethoxyphenyl)-1H-indazol-5-yl]propanamide, Ribosomal protein S6 kinase alpha-3 | | Authors: | Miller, R.M, Paavilainen, V.O, Krishnan, S, Serafimova, I.M, Taunton, J. | | Deposit date: | 2013-02-28 | | Release date: | 2013-04-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1002 Å) | | Cite: | Electrophilic fragment-based design of reversible covalent kinase inhibitors.
J.Am.Chem.Soc., 135, 2013
|
|
8S9W
 
 | | Murine S100A7/S100A15 in presence of calcium | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Harrison, S.A, Naretto, A, Balakrishnan, S, Chazin, W.J. | | Deposit date: | 2023-03-30 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Comparative analysis of the physical properties of murine and human S100A7: Insight into why zinc piracy is mediated by human but not murine S100A7.
J.Biol.Chem., 299, 2023
|
|
4MAO
 
 | | RSK2 T493M C-Terminal Kinase Domain in Complex with RMM58 | | Descriptor: | (2Z)-2-(1H-1,2,4-triazol-1-yl)-3-[3-(3,4,5-trimethoxyphenyl)-1H-indazol-5-yl]prop-2-enenitrile, Ribosomal protein S6 kinase alpha-3, SODIUM ION | | Authors: | Miller, R.M, Taunton, J. | | Deposit date: | 2013-08-16 | | Release date: | 2014-10-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design of reversible, cysteine-targeted Michael acceptors guided by kinetic and computational analysis.
J.Am.Chem.Soc., 136, 2014
|
|
6VWV
 
 | |
6VWT
 
 | |
4GJY
 
 | | JMJD5 in complex with N-Oxalylglycine | | Descriptor: | COBALT (II) ION, JmjC domain-containing protein 5, N-OXALYLGLYCINE | | Authors: | Del Rizzo, P.A, Trievel, R.C. | | Deposit date: | 2012-08-10 | | Release date: | 2012-09-05 | | Last modified: | 2012-11-14 | | Method: | X-RAY DIFFRACTION (1.2492 Å) | | Cite: | Crystal Structure and Functional Analysis of JMJD5 Indicate an Alternate Specificity and Function.
Mol.Cell.Biol., 32, 2012
|
|
6OCE
 
 | | Structure of the rice hyperosmolality-gated ion channel OSCA1.2 | | Descriptor: | stress-gated cation channel 1.2 | | Authors: | Maity, K, Heumann, J.M, McGrath, A.P, Chang, G, Stowell, M.H. | | Deposit date: | 2019-03-23 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Cryo-EM structure of OSCA1.2 fromOryza sativaelucidates the mechanical basis of potential membrane hyperosmolality gating.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7Y0D
 
 | |
8P4X
 
 | | FAD_ox bound dark state structure of PdLCry | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, Putative light-receptive cryptochrome (Fragment) | | Authors: | Behrmann, E, Behrmann, H. | | Deposit date: | 2023-05-23 | | Release date: | 2023-11-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | A marine cryptochrome with an inverse photo-oligomerization mechanism.
Nat Commun, 14, 2023
|
|
4GJZ
 
 | | JMJD5 in complex with 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, BETA-MERCAPTOETHANOL, COBALT (II) ION, ... | | Authors: | Del Rizzo, P.A, Trievel, R.C. | | Deposit date: | 2012-08-10 | | Release date: | 2012-09-05 | | Last modified: | 2012-11-14 | | Method: | X-RAY DIFFRACTION (1.0481 Å) | | Cite: | Crystal Structure and Functional Analysis of JMJD5 Indicate an Alternate Specificity and Function.
Mol.Cell.Biol., 32, 2012
|
|
1S6P
 
 | | CRYSTAL STRUCTURE OF HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R100943 | | Descriptor: | 1-(4-CYANO-PHENYL)-3-[2-(2,6-DICHLORO-PHENYL)-1-IMINO-ETHYL]-THIOUREA, MAGNESIUM ION, POL polyprotein [Contains: Reverse transcriptase] | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2004-01-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants
J.Med.Chem., 47, 2004
|
|
4M8T
 
 | |
4LV0
 
 | |
4LV2
 
 | |
4LV1
 
 | |
