1WRF
 
 | | Refined solution structure of Der f 2, The Major Mite Allergen from Dermatophagoides farinae | | Descriptor: | Mite group 2 allergen Der f 2 | | Authors: | Ichikawa, S, Takai, T, Inoue, T, Yuuki, T, Okumura, Y, Ogura, K, Inagaki, F, Hatanaka, H. | | Deposit date: | 2004-10-15 | | Release date: | 2005-04-19 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | NMR Study on the Major Mite Allergen Der f 2: Its Refined Tertiary Structure, Epitopes for Monoclonal Antibodies and Characteristics Shared by ML Protein Group Members
J.Biochem.(Tokyo), 137, 2005
|
|
1AHM
 
 | | DER F 2, THE MAJOR MITE ALLERGEN FROM DERMATOPHAGOIDES FARINAE, NMR, 10 STRUCTURES | | Descriptor: | DER F 2 | | Authors: | Ichikawa, S, Hatanaka, H, Yuuki, T, Iwamoto, N, Ogura, K, Okumura, Y, Inagaki, F. | | Deposit date: | 1997-04-07 | | Release date: | 1998-04-08 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Der f 2, the major mite allergen for atopic diseases.
J.Biol.Chem., 273, 1998
|
|
1AHK
 
 | | DER F 2, THE MAJOR MITE ALLERGEN FROM DERMATOPHAGOIDES FARINAE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DER F 2 | | Authors: | Ichikawa, S, Hatanaka, H, Yuuki, T, Iwamoto, N, Ogura, K, Okumura, Y, Inagaki, F. | | Deposit date: | 1997-04-07 | | Release date: | 1998-04-08 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Der f 2, the major mite allergen for atopic diseases.
J.Biol.Chem., 273, 1998
|
|
5CKR
 
 | | Crystal Structure of MraY in complex with Muraymycin D2 | | Descriptor: | Muraymycin D2, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Lee, S.Y, Chung, B.C, Mashalidis, E.H, Tanino, T, Kim, M, Hong, J, Ichikawa, S. | | Deposit date: | 2015-07-15 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural insights into inhibition of lipid I production in bacterial cell wall synthesis.
Nature, 533, 2016
|
|
2RRR
 
 | | DNA oligomer containing ethylene cross-linked cyclic 2'-deoxyuridylate dimer | | Descriptor: | DNA (5'-D(*CP*CP*TP*TP*CP*AP*TP*TP*AP*CP*AP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*GP*TP*AP*AP*TP*GP*AP*AP*GP*G)-3') | | Authors: | Furuita, K, Murata, S, Jee, J.G, Ichikawa, S, Matsuda, A, Kojima, C. | | Deposit date: | 2011-03-24 | | Release date: | 2011-06-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Feature of Bent DNA Recognized by HMGB1
J.Am.Chem.Soc., 133, 2011
|
|
2RRQ
 
 | | DNA oligomer containing propylene cross-linked cyclic 2'-deoxyuridylate dimer | | Descriptor: | DNA (5'-D(*CP*CP*TP*TP*CP*AP*(JDT)P*TP*AP*CP*AP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*GP*TP*AP*AP*TP*GP*AP*AP*GP*G)-3') | | Authors: | Furuita, K, Murata, S, Jee, J.G, Ichikawa, S, Matsuda, A, Kojima, C. | | Deposit date: | 2011-03-24 | | Release date: | 2011-06-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Feature of Bent DNA Recognized by HMGB1
J.Am.Chem.Soc., 133, 2011
|
|
1AK7
 
 | | DESTRIN, NMR, 20 STRUCTURES | | Descriptor: | DESTRIN | | Authors: | Hatanaka, H, Moriyama, K, Ogura, K, Ichikawa, S, Yahara, I, Inagaki, F. | | Deposit date: | 1997-05-29 | | Release date: | 1997-10-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Tertiary structure of destrin and structural similarity between two actin-regulating protein families.
Cell(Cambridge,Mass.), 85, 1996
|
|
1AK6
 
 | | DESTRIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DESTRIN | | Authors: | Hatanaka, H, Moriyama, K, Ogura, K, Ichikawa, S, Yahara, I, Inagaki, F. | | Deposit date: | 1997-05-29 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Tertiary structure of destrin and structural similarity between two actin-regulating protein families.
Cell(Cambridge,Mass.), 85, 1996
|
|
1HRE
 
 | | SOLUTION STRUCTURE OF THE EPIDERMAL GROWTH FACTOR-LIKE DOMAIN OF HEREGULIN-ALPHA, A LIGAND FOR P180ERB4 | | Descriptor: | HEREGULIN ALPHA | | Authors: | Nagata, K, Kohda, D, Hatanaka, H, Ichikawa, S, Inagaki, F. | | Deposit date: | 1994-07-21 | | Release date: | 1994-10-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the epidermal growth factor-like domain of heregulin-alpha, a ligand for p180erbB-4.
EMBO J., 13, 1994
|
|
1HRF
 
 | | SOLUTION STRUCTURE OF THE EPIDERMAL GROWTH FACTOR-LIKE DOMAIN OF HEREGULIN-ALPHA, A LIGAND FOR P180ERB4 | | Descriptor: | HEREGULIN ALPHA | | Authors: | Nagata, K, Kohda, D, Hatanaka, H, Ichikawa, S, Inagaki, F. | | Deposit date: | 1994-07-21 | | Release date: | 1994-10-15 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the epidermal growth factor-like domain of heregulin-alpha, a ligand for p180erbB-4.
EMBO J., 13, 1994
|
|
1IP9
 
 | | SOLUTION STRUCTURE OF THE PB1 DOMAIN OF BEM1P | | Descriptor: | BEM1 PROTEIN | | Authors: | Terasawa, H, Noda, Y, Ito, T, Hatanaka, H, Ichikawa, S, Ogura, K, Sumimoto, H, Inagaki, F. | | Deposit date: | 2001-04-26 | | Release date: | 2001-08-15 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and ligand recognition of the PB1 domain: a novel protein module binding to the PC motif.
EMBO J., 20, 2001
|
|
1IPG
 
 | | SOLUTION STRUCTURE OF THE PB1 DOMAIN OF BEM1P | | Descriptor: | BEM1 PROTEIN | | Authors: | Terasawa, H, Noda, Y, Ito, T, Hatanaka, H, Ichikawa, S, Ogura, K, Sumimoto, H, Inagaki, F. | | Deposit date: | 2001-05-14 | | Release date: | 2001-08-15 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and ligand recognition of the PB1 domain: a novel protein module binding to the PC motif.
EMBO J., 20, 2001
|
|
8CXR
 
 | | Crystal structure of MraY bound to a sphaerimicin analogue | | Descriptor: | (1S,4R,5S,6R,7S,9S,10S,11S,13S,14R)-9-[(2S,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxyoxolan-2-yl]-14-(hexadecanoyloxy)-5,6,13-trihydroxy-8,16-dioxa-2,11-diazatricyclo[9.3.1.1~4,7~]hexadecane-10-carboxylic acid, MraYAA nanobody, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Mashalidis, E.H, Lee, S.Y. | | Deposit date: | 2022-05-22 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Synthesis of macrocyclic nucleoside antibacterials and their interactions with MraY.
Nat Commun, 13, 2022
|
|
9B70
 
 | | Cryo-EM structure of MraY in complex with analogue 2 | | Descriptor: | (2~{S},3~{S})-3-[(2~{S},3~{R},4~{S},5~{R})-5-(aminomethyl)-3,4-bis(oxidanyl)oxolan-2-yl]oxy-2-[[3-[[[(2~{S})-6-azanyl-2-(hexadecanoylamino)hexanoyl]amino]methyl]phenyl]methylamino]-3-[(2~{S},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]propanoic acid, MraYAA nanobody, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Hao, A, Lee, S.-Y. | | Deposit date: | 2024-03-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Development of a natural product optimization strategy for inhibitors against MraY, a promising antibacterial target.
Nat Commun, 15, 2024
|
|
9B71
 
 | | Cryo-EM structure of MraY in complex with analogue 3 | | Descriptor: | (2~{S},3~{S})-3-[(2~{S},3~{R},4~{S},5~{R})-5-(aminomethyl)-3,4-bis(oxidanyl)oxolan-2-yl]oxy-3-[(2~{S},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]-2-[[4-[[[(2~{S})-5-carbamimidamido-2-(hexadecanoylamino)pentanoyl]amino]methyl]phenyl]methylamino]propanoic acid, MraYAA Nanobody, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Hao, A, Lee, S.-Y. | | Deposit date: | 2024-03-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Development of a natural product optimization strategy for inhibitors against MraY, a promising antibacterial target.
Nat Commun, 15, 2024
|
|
6BW5
 
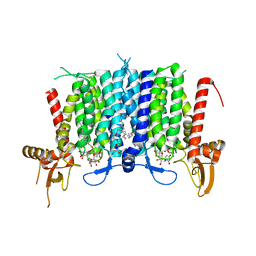 | | Human GPT (DPAGT1) in complex with tunicamycin | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Tunicamycin, UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase | | Authors: | Yoo, J, Kuk, A.C.Y, Mashalidis, E.H, Lee, S.-Y. | | Deposit date: | 2017-12-14 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | GlcNAc-1-P-transferase-tunicamycin complex structure reveals basis for inhibition of N-glycosylation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6BW6
 
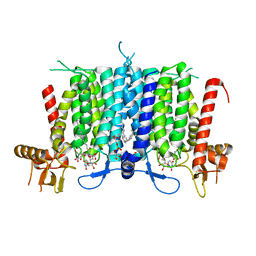 | | Human GPT (DPAGT1) H129 variant in complex with tunicamycin | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Tunicamycin, UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase | | Authors: | Yoo, J, Kuk, A.C.Y, Mashalidis, E.H, Lee, S.-Y. | | Deposit date: | 2017-12-14 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | GlcNAc-1-P-transferase-tunicamycin complex structure reveals basis for inhibition of N-glycosylation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6OYH
 
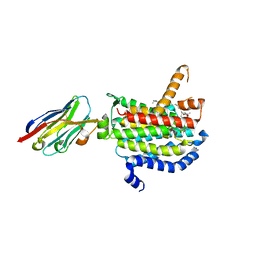 | | Crystal structure of MraY bound to carbacaprazamycin | | Descriptor: | (5S)-5'-O-(5-amino-5-deoxy-beta-D-ribofuranosyl)-5'-C-[(2S,5S,6S)-5-carboxy-6-heptadecyl-1,4-dimethyl-3-oxo-1,4-diazepan-2-yl]uridine, MraYAA nanobody, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Mashalidis, E.H, Lee, S.Y. | | Deposit date: | 2019-05-14 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Chemical logic of MraY inhibition by antibacterial nucleoside natural products.
Nat Commun, 10, 2019
|
|
6OYZ
 
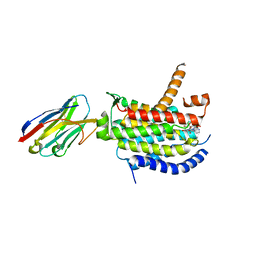 | | Crystal structure of MraY bound to capuramycin | | Descriptor: | (2~{S},3~{S},4~{S})-2-[(1~{R})-2-azanyl-1-[(2~{S},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3-methoxy-4-oxidanyl-oxolan-2-yl]-2-oxidanylidene-ethoxy]-3,4-bis(oxidanyl)-~{N}-[(3~{S})-2-oxidanylideneazepan-3-yl]-3,4-dihydro-2~{H}-pyran-6-carboxamide, MraYAA nanobody, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Mashalidis, E.H, Lee, S.Y. | | Deposit date: | 2019-05-15 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Chemical logic of MraY inhibition by antibacterial nucleoside natural products.
Nat Commun, 10, 2019
|
|
6OZ6
 
 | | Crystal structure of MraY bound to 3'-hydroxymureidomycin A | | Descriptor: | (2~{S})-2-[[(2~{S})-1-[[(2~{S},3~{S})-3-[[(2~{S})-2-azanyl-3-(3-hydroxyphenyl)propanoyl]-methyl-amino]-1-[[(~{Z})-[(3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-ylidene]methyl]amino]-1-oxidanylidene-butan-2-yl]amino]-4-methylsulfanyl-1-oxidanylidene-butan-2-yl]carbamoylamino]-3-(3-hydroxyphenyl)propanoic acid, MraYAA nanobody, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Mashalidis, E.H, Lee, S.Y. | | Deposit date: | 2019-05-15 | | Release date: | 2019-07-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Chemical logic of MraY inhibition by antibacterial nucleoside natural products.
Nat Commun, 10, 2019
|
|
7V4M
 
 | |
7V4O
 
 | | Unique non-heme hydroxylase in biosynthesis of nucleoside antibiotic pathway uncover mechanism of reaction | | Descriptor: | 3,6,9,12,15-PENTAOXAHEPTADECANE, Beta-hydroxylase | | Authors: | Li, T.L, Saeid, M.Z. | | Deposit date: | 2021-08-13 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | beta-Hydroxylation of alpha-amino-beta-hydroxylbutanoyl-glycyluridine catalyzed by a nonheme hydroxylase ensures the maturation of caprazamycin
Commun Chem, 5, 2022
|
|
7V4N
 
 | |
7V4F
 
 | |
7V4P
 
 | |
