2VTY
 
 | | Vaccinia virus anti-apoptotic F1L is a novel Bcl-2-like domain swapped dimer | | Descriptor: | PROTEIN F1 | | Authors: | Kvansakul, M, Yang, H, Fairlie, W.D, Czabotar, P.E, Fischer, S.F, Perugini, M.A, Huang, D.C.S, Colman, P.M. | | Deposit date: | 2008-05-19 | | Release date: | 2008-06-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Vaccinia Virus Anti-Apoptotic F1L is a Novel Bcl-2-Like Domain-Swapped Dimer that Binds a Highly Selective Subset of Bh3-Containing Death Ligands.
Cell Death Differ., 15, 2008
|
|
2XPX
 
 | | Crystal structure of BHRF1:Bak BH3 complex | | Descriptor: | 1,2-ETHANEDIOL, APOPTOSIS REGULATOR BHRF1, BCL-2 HOMOLOGOUS ANTAGONIST/KILLER, ... | | Authors: | Kvansakul, M, Huang, D.C.S, Colman, P.M. | | Deposit date: | 2010-08-31 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis for Apoptosis Inhibition by Epstein-Barr Virus Bhrf1.
Plos Pathog., 6, 2010
|
|
1O0L
 
 | | THE STRUCTURE OF BCL-W REVEALS A ROLE FOR THE C-TERMINAL RESIDUES IN MODULATING BIOLOGICAL ACTIVITY | | Descriptor: | Apoptosis regulator Bcl-W | | Authors: | Hinds, M.G, Lackmann, M, Skea, G.L, Harrison, P.J, Huang, D.C.S, Day, C.L. | | Deposit date: | 2003-02-22 | | Release date: | 2003-04-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of Bcl-w reveals a role for the C-terminal residues in modulating biological activity
Embo J., 22, 2003
|
|
2JBX
 
 | | Crystal Structure of the myxoma virus anti-apoptotic protein M11L | | Descriptor: | M11L PROTEIN | | Authors: | Kvansakul, M, Van Delft, M.F, Lee, E.F, Gulbis, J.M, Fairlie, W.D, Huang, D.C.S, Colman, P.M. | | Deposit date: | 2006-12-14 | | Release date: | 2007-03-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | A Structural Viral Mimic of Prosurvival Bcl-2: A Pivotal Role for Sequestering Proapoptotic Bax and Bak.
Mol.Cell, 25, 2007
|
|
2JM6
 
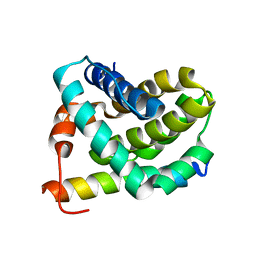 | | Solution structure of MCL-1 complexed with NOXAB | | Descriptor: | Myeloid cell leukemia-1 protein Mcl-1 homolog, Noxa | | Authors: | Czabotar, P.E, Lee, E.F, van Delft, M.F, Day, C.L, Smith, B.J, Huang, D.C.S, Fairlie, W.D, Hinds, M.G, Colman, P.M. | | Deposit date: | 2006-10-17 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the degradation of Mcl-1 induced by BH3 domains
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2WH6
 
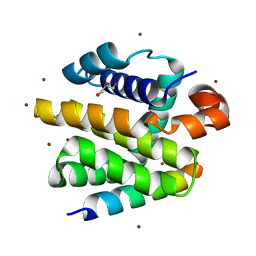 | | Crystal structure of anti-apoptotic BHRF1 in complex with the Bim BH3 domain | | Descriptor: | BCL-2-LIKE PROTEIN 11, BROMIDE ION, EARLY ANTIGEN PROTEIN R, ... | | Authors: | Kvansakul, M, Huang, D.C.S, Colman, P.M. | | Deposit date: | 2009-05-01 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for apoptosis inhibition by Epstein-Barr virus BHRF1.
PLoS Pathog., 6, 2010
|
|
2YQ7
 
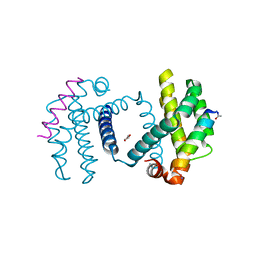 | | Structure of Bcl-xL bound to BimLOCK | | Descriptor: | BCL-2-LIKE PROTEIN 1, BCL-2-LIKE PROTEIN 11, GLYCEROL | | Authors: | Smith, B.J, Czabotar, P.E. | | Deposit date: | 2012-11-06 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Stabilizing the Pro-Apoptotic Bimbh3 Helix (Bimsahb) Does not Necessarily Enhance Affinity or Biological Activity.
Acs Chem.Biol., 8, 2013
|
|
2YQ6
 
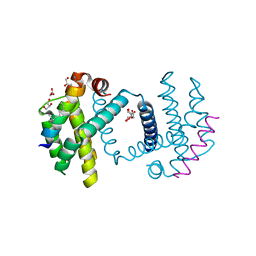 | | Structure of Bcl-xL bound to BimSAHB | | Descriptor: | BCL-2-LIKE PROTEIN 1, BIM BETA 5, GLYCEROL | | Authors: | Smith, B.J, Czabotar, P.E. | | Deposit date: | 2012-11-06 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Stabilizing the Pro-Apoptotic Bimbh3 Helix (Bimsahb) Does not Necessarily Enhance Affinity or Biological Activity.
Acs Chem.Biol., 8, 2013
|
|
5U6V
 
 | | X-ray crystal structure of 1,2,3-triazolobenzodiazepine in complex with BRD2(D2) | | Descriptor: | 1,2-ETHANEDIOL, 5-[7-(4-chlorophenyl)-1-methyl-6,7-dihydro-5H-[1,2,3]triazolo[1,5-d][1,4]benzodiazepin-9-yl]pyridin-2-amine, Bromodomain-containing protein 2 | | Authors: | Hatfaludi, T, Sharp, P.P, Garnier, J.-M, Burns, C.J, Czabotar, P.E. | | Deposit date: | 2016-12-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.775 Å) | | Cite: | Design, Synthesis, and Biological Activity of 1,2,3-Triazolobenzodiazepine BET Bromodomain Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
3D7V
 
 | | Crystal structure of Mcl-1 in complex with an Mcl-1 selective BH3 ligand | | Descriptor: | Bcl-2-like protein 11, Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION | | Authors: | Lee, E.F, Czabotar, P.E, Colman, P.M, Fairlie, W.D. | | Deposit date: | 2008-05-22 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A novel BH3 ligand that selectively targets Mcl-1 reveals that apoptosis can proceed without Mcl-1 degradation.
J.Cell Biol., 180, 2008
|
|
2V6Q
 
 | |
6MCY
 
 | | Crystal structure of mouse Bak | | Descriptor: | 1,2-ETHANEDIOL, Bcl-2 homologous antagonist/killer, FORMIC ACID | | Authors: | Brouwer, J.M, Czabotar, P.E, Colman, P.M. | | Deposit date: | 2018-09-03 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.748 Å) | | Cite: | A small molecule interacts with VDAC2 to block mouse BAK-driven apoptosis.
Nat.Chem.Biol., 15, 2019
|
|
4B4S
 
 | |
6O0L
 
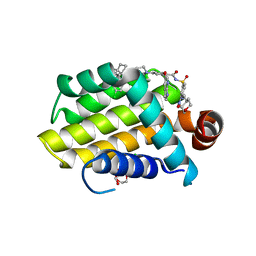 | | crystal structure of BCL-2 G101V mutation with venetoclax | | Descriptor: | 4-{4-[(4'-chloro-5,5-dimethyl[3,4,5,6-tetrahydro[1,1'-biphenyl]]-2-yl)methyl]piperazin-1-yl}-N-[(3-nitro-4-{[(oxan-4-yl )methyl]amino}phenyl)sulfonyl]-2-[(1H-pyrrolo[2,3-b]pyridin-5-yl)oxy]benzamide, Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2, CHLORIDE ION, ... | | Authors: | Birkinshaw, R.W, Luo, C.S, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-02-16 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of BCL-2 in complex with venetoclax reveal the molecular basis of resistance mutations.
Nat Commun, 10, 2019
|
|
6O0P
 
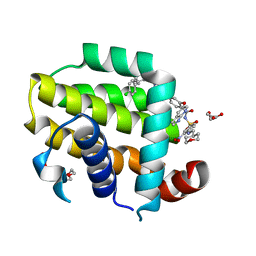 | | crystal structure of BCL-2 G101A mutation with venetoclax | | Descriptor: | 4-{4-[(4'-chloro-5,5-dimethyl[3,4,5,6-tetrahydro[1,1'-biphenyl]]-2-yl)methyl]piperazin-1-yl}-N-[(3-nitro-4-{[(oxan-4-yl )methyl]amino}phenyl)sulfonyl]-2-[(1H-pyrrolo[2,3-b]pyridin-5-yl)oxy]benzamide, Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2, DI(HYDROXYETHYL)ETHER | | Authors: | Birkinshaw, R.W, Luo, C.S, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-02-17 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of BCL-2 in complex with venetoclax reveal the molecular basis of resistance mutations.
Nat Commun, 10, 2019
|
|
6O0M
 
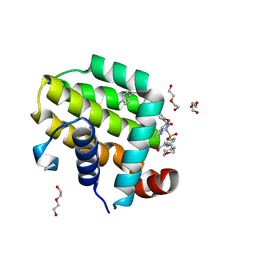 | | crystal structure of BCL-2 F104L mutation with venetoclax | | Descriptor: | 4-{4-[(4'-chloro-5,5-dimethyl[3,4,5,6-tetrahydro[1,1'-biphenyl]]-2-yl)methyl]piperazin-1-yl}-N-[(3-nitro-4-{[(oxan-4-yl )methyl]amino}phenyl)sulfonyl]-2-[(1H-pyrrolo[2,3-b]pyridin-5-yl)oxy]benzamide, Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2, DI(HYDROXYETHYL)ETHER | | Authors: | Birkinshaw, R.W, Luo, C.S, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-02-16 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of BCL-2 in complex with venetoclax reveal the molecular basis of resistance mutations.
Nat Commun, 10, 2019
|
|
6O0K
 
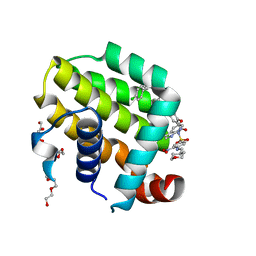 | | crystal structure of BCL-2 with venetoclax | | Descriptor: | 4-{4-[(4'-chloro-5,5-dimethyl[3,4,5,6-tetrahydro[1,1'-biphenyl]]-2-yl)methyl]piperazin-1-yl}-N-[(3-nitro-4-{[(oxan-4-yl )methyl]amino}phenyl)sulfonyl]-2-[(1H-pyrrolo[2,3-b]pyridin-5-yl)oxy]benzamide, Apoptosis regulator Bcl-2, NONAETHYLENE GLYCOL | | Authors: | Birkinshaw, R.W, Luo, C.S, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-02-16 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structures of BCL-2 in complex with venetoclax reveal the molecular basis of resistance mutations.
Nat Commun, 10, 2019
|
|
4C5D
 
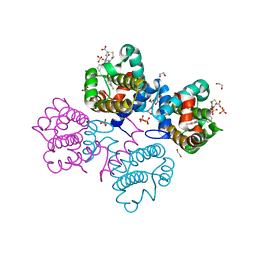 | | Crystal structure of Bcl-xL in complex with benzoylurea compound (42) | | Descriptor: | (R)-3-(4-BROMOBENZYLTHIO)-2-(3-(3-((2,4-DIFLUOROPHENYL)ETHYNYL)BENZOYL)-3-PROPYLUREIDO)PROPANOIC ACID, 1,2-ETHANEDIOL, BCL-2-LIKE PROTEIN 1, ... | | Authors: | Roy, M.J, Brady, R.M, Lessene, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2013-09-11 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | De-Novo Designed Library of Benzoylureas as Inhibitors of Bcl-Xl: Synthesis, Structural and Biochemical Characterization.
J.Med.Chem., 57, 2014
|
|
6O0O
 
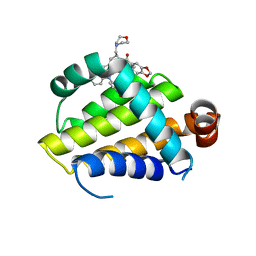 | | crystal structure of BCL-2 G101V mutation with S55746 | | Descriptor: | Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2, ~{N}-(4-hydroxyphenyl)-3-[6-[[(3~{S})-3-(morpholin-4-ylmethyl)-3,4-dihydro-1~{H}-isoquinolin-2-yl]carbonyl]-1,3-benzodioxol-5-yl]-~{N}-phenyl-5,6,7,8-tetrahydroindolizine-1-carboxamide | | Authors: | Birkinshaw, R.W, Luo, C.S, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-02-17 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structures of BCL-2 in complex with venetoclax reveal the molecular basis of resistance mutations.
Nat Commun, 10, 2019
|
|
4C52
 
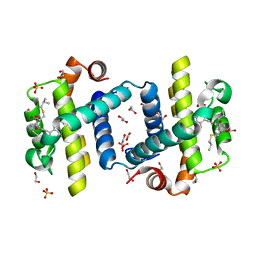 | | Crystal structure of Bcl-xL in complex with benzoylurea compound (39b) | | Descriptor: | (R)-2-(3-(3-((2,4-DIFLUOROPENYL)ETHYNYL)BENZOYL)-3-PROPYLUREIDO)-3-(ISOBUTYLTHIO) PROPANOIC ACID, 1,2-ETHANEDIOL, BCL-2-LIKE PROTEIN 1, ... | | Authors: | Roy, M.J, Brady, R.M, Lessene, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2013-09-10 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | De-Novo Designed Library of Benzoylureas as Inhibitors of Bcl-Xl: Synthesis, Structural and Biochemical Characterization.
J.Med.Chem., 57, 2014
|
|
6O5Z
 
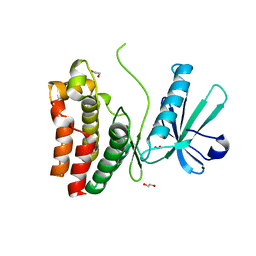 | | Crystal Structure of the human MLKL pseudokinase domain bound to compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 1-[2-fluoranyl-5-(trifluoromethyl)phenyl]-3-[4-[methyl-[2-[(3-sulfamoylphenyl)amino]pyrimidin-4-yl]amino]phenyl]urea, Mixed lineage kinase domain-like protein | | Authors: | Cowan, A.D, Murphy, J.M, Pierotti, C.L, Lessene, G.L, Czabotar, P.E. | | Deposit date: | 2019-03-04 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | Potent Inhibition of Necroptosis by Simultaneously Targeting Multiple Effectors of the Pathway.
Acs Chem.Biol., 15, 2020
|
|
3ZLO
 
 | | Crystal structure of BCL-XL in complex with inhibitor (Compound 6) | | Descriptor: | 2-[(8E)-8-(1,3-benzothiazol-2-ylhydrazinylidene)-6,7-dihydro-5H-naphthalen-2-yl]-5-(4-phenylbutyl)-1,3-thiazole-4-carboxylic acid, BCL-2-LIKE PROTEIN 1 | | Authors: | Czabotar, P.E, Lessene, G.L, Smith, B.J, Colman, P.M. | | Deposit date: | 2013-02-04 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structure-Guided Design of a Selective Bcl-Xl Inhibitor
Nat.Chem.Biol., 9, 2013
|
|
3ZLR
 
 | | Crystal structure of BCL-XL in complex with inhibitor (WEHI-539) | | Descriptor: | 1,2-ETHANEDIOL, 5-[3-[4-(aminomethyl)phenoxy]propyl]-2-[(8E)-8-(1,3-benzothiazol-2-ylhydrazinylidene)-6,7-dihydro-5H-naphthalen-2-yl]-1,3-thiazole-4-carboxylic acid, BCL-2-LIKE PROTEIN 1, ... | | Authors: | Czabotar, P.E, Lessene, G.L, Smith, B.J, Colman, P.M. | | Deposit date: | 2013-02-04 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.026 Å) | | Cite: | Structure-Guided Design of a Selective Bcl-Xl Inhibitor
Nat.Chem.Biol., 9, 2013
|
|
3ZLN
 
 | | Crystal structure of BCL-XL in complex with inhibitor (Compound 3) | | Descriptor: | 1,2-ETHANEDIOL, 6-[(8E)-8-(1,3-benzothiazol-2-ylhydrazinylidene)-6,7-dihydro-5H-naphthalen-2-yl]pyridine-2-carboxylic acid, BCL-2-LIKE PROTEIN 1, ... | | Authors: | Czabotar, P.E, Lessene, G.L, Smith, B.J, Colman, P.M. | | Deposit date: | 2013-02-04 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.288 Å) | | Cite: | Structure-Guided Design of a Selective Bcl-Xl Inhibitor
Nat.Chem.Biol., 9, 2013
|
|
3ZK6
 
 | | Crystal structure of Bcl-xL in complex with inhibitor (Compound 2). | | Descriptor: | BCL-2-LIKE PROTEIN 1, N-(3-(5-(1-(2-(benzo[d]thiazol-2-yl)hydrazono)ethyl)furan-2-yl)phenylsulfonyl)-6-phenylhexanamide | | Authors: | Czabotar, P.E, Lessene, G.L, Smith, B.J, Colman, P.M. | | Deposit date: | 2013-01-22 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure-Guided Design of a Selective Bcl-Xl Inhibitor
Nat.Chem.Biol., 9, 2013
|
|
