3K12
 
 | | Crystal structure of an uncharacterized protein A6V7T0 from Pseudomonas aeruginosa | | Descriptor: | GLYCEROL, uncharacterized protein A6V7T0 | | Authors: | Ho, M, Ramagopal, U.A, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-25 | | Release date: | 2009-10-27 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structure of an uncharacterized protein A6V7T0 from Pseudomonas aeruginosa
To be published
|
|
3K8Q
 
 | | Crystal structure of human purine nucleoside phosphorylase in complex with SerMe-Immucillin H | | Descriptor: | 7-({[2-hydroxy-1-(hydroxymethyl)ethyl]amino}methyl)-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Ho, M, Almo, S.C, Scharmm, V.L. | | Deposit date: | 2009-10-14 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human nucleoside phosphorylase in complex with SerMe-ImmH
Proc.Natl.Acad.Sci.USA, 2010
|
|
3K8O
 
 | | Crystal structure of human purine nucleoside phosphorylase in complex with DATMe-ImmH | | Descriptor: | 7-({[(1R,2S)-2,3-DIHYDROXY-1-(HYDROXYMETHYL)PROPYL]AMINO}METHYL)-3,5-DIHYDRO-4H-PYRROLO[3,2-D]PYRIMIDIN-4-ONE, Purine nucleoside phosphorylase, SULFATE ION | | Authors: | Ho, M, Rinaldo-matthis, A, Almo, S.C, Schramm, V.L. | | Deposit date: | 2009-10-14 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human purine nucleoside phosphorylase in complex with DATMe-Immucillin H
Proc.Natl.Acad.Sci.USA, 2010
|
|
3OZE
 
 | |
3OZD
 
 | | Crystal Structure of human 5'-deoxy-5'-methyladenosine phosphorylase in complex with pCl-phenylthioDADMeImmA | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-{[(4-chlorophenyl)sulfanyl]methyl}pyrrolidin-3-ol, S-methyl-5'-thioadenosine phosphorylase | | Authors: | Ho, M, Guan, R, Almo, S.C, Schramm, V.L. | | Deposit date: | 2010-09-24 | | Release date: | 2011-09-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of human 5'-deoxy-5'-methyladenosine phosphorylase
to be published
|
|
3OZC
 
 | | Crystal Structure of human 5'-deoxy-5'-methyladenosine phosphorylase in complex with pCl-phenylthioDADMeImmA | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-{[(4-chlorophenyl)sulfanyl]methyl}pyrrolidin-3-ol, PHOSPHATE ION, S-methyl-5'-thioadenosine phosphorylase | | Authors: | Ho, M, Guan, R, Almo, S.C, Schramm, V.L. | | Deposit date: | 2010-09-24 | | Release date: | 2011-09-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure of human 5'-deoxy-5'-methyladenosine phosphorylase
to be published
|
|
3PHB
 
 | | Crystal Structure of human purine nucleoside phosphorylase in complex with DADMe-ImmG | | Descriptor: | 2-amino-7-{[(3R,4R)-3-hydroxy-4-(hydroxymethyl)pyrrolidin-1-yl]methyl}-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Ho, M, Cassera, M.B, Murkin, A.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2010-11-03 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of human purine nucleoside phosphorylase in complex with DADMe-ImmG
to be published
|
|
3PHC
 
 | | Crystal Structure of Plasmodium falciparum purine nucleoside phosphorylase in complex with DADMe-ImmG | | Descriptor: | 2-amino-7-{[(3R,4R)-3-hydroxy-4-(hydroxymethyl)pyrrolidin-1-yl]methyl}-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Ho, M, Edwards, A.A, Almo, S.C, Schramm, V.L. | | Deposit date: | 2010-11-03 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Plasmodium falciparum purine nucleoside phosphorylase in complex with DADMe-ImmG
To be Published
|
|
3D0O
 
 | |
3EEI
 
 | | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from neisseria meningitidis in complex with methylthio-immucillin-A | | Descriptor: | (3S,4R)-2-(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-5-[(METHYLSULFANYL)METHYL]PYRROLIDINE-3,4-DIOL, 5-methylthioadenosine nucleosidase/S-adenosylhomocysteine nucleosidase | | Authors: | Ho, M, Rinaldo-matthis, A, Brown, R.L, Norris, G.E, Tyler, P.C, Furneaux, R.H, Almo, S.C, Schramm, V.L. | | Deposit date: | 2008-09-04 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from neisseria meningitidis in complex with methylthio-immucillin-A
To be Published
|
|
3CHG
 
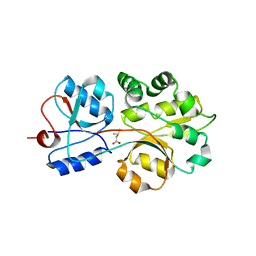 | | The compatible solute-binding protein OpuAC from Bacillus subtilis in complex with DMSA | | Descriptor: | (dimethyl-lambda~4~-sulfanyl)acetic acid, Glycine betaine-binding protein | | Authors: | Smits, S.H.J, Hoing, M, Lecher, J, Jebbar, M, Schmitt, L, Bremer, E. | | Deposit date: | 2008-03-09 | | Release date: | 2008-08-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Compatible-Solute-Binding Protein OpuAC from Bacillus subtilis: Ligand Binding, Site-Directed Mutagenesis, and Crystallographic Studies
J.Bacteriol., 190, 2008
|
|
7T62
 
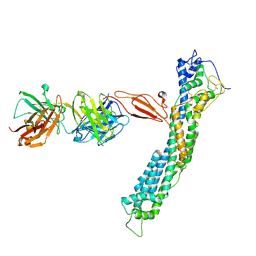 | | GPC2 HEP CT3 complex | | Descriptor: | CT3, Glypican-2 | | Authors: | Zhu, J, Cachau, R, De Val Alda, N, Li, N, Ho, M. | | Deposit date: | 2021-12-13 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (21 Å) | | Cite: | CAR T cells targeting tumor-associated exons of glypican 2 regress neuroblastoma in mice.
Cell Rep Med, 2, 2021
|
|
7TPR
 
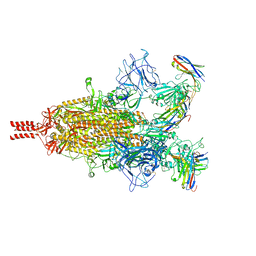 | | Camel nanobodies 7A3 and 8A2 broadly neutralize SARS-CoV-2 variants | | Descriptor: | Nanobody 7A3, Nanobody 8A2, Spike glycoprotein | | Authors: | Butay, K.J, Zhu, J, Dandey, V.P, Hong, J, Kwon, H.J, Chen, C.Z, Duan, Z, Li, D, Ren, H, Liang, T, Martin, N, Esposito, D, Ortega-Rodriguez, U, Xu, M, Xie, H, Ho, M, Cachau, R, Borgnia, M.J. | | Deposit date: | 2022-01-25 | | Release date: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Camel nanobodies broadly neutralize SARS-CoV-2 variants
bioRxiv, 2021
|
|
3LMD
 
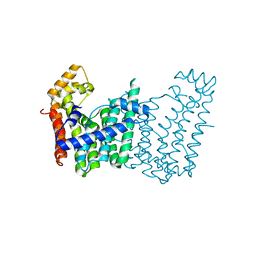 | | Crystal structure of geranylgeranyl pyrophosphate synthase from corynebacterium glutamicum atcc 13032 | | Descriptor: | Geranylgeranyl pyrophosphate synthase | | Authors: | Patskovsky, Y, Ho, M, Toro, R, Rutter, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-01-29 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Geranylgeranyl Pyrophosphate Synthase from Corynebacterium Glutamicum
To be Published
|
|
3D4P
 
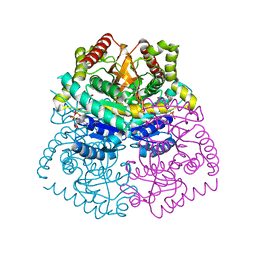 | |
2N9W
 
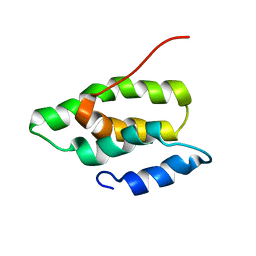 | | Solution NMR Structure of the membrane localization domain from the Ras/Rap1-specific endopeptidase (RRSP) of the Vibrio vulnificus multifunctional autoprocessing repeats-in-toxins (MARTX) toxin | | Descriptor: | RTX toxin | | Authors: | Hisao, G.S, Brothers, M.C, Ho, M, Wilson, B.A, Rienstra, C.M. | | Deposit date: | 2015-12-12 | | Release date: | 2016-12-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The membrane localization domains of two distinct bacterial toxins form a 4-helix-bundle in solution.
Protein Sci., 26, 2017
|
|
2N9V
 
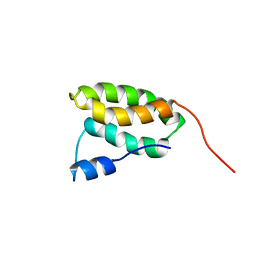 | | Solution NMR Structure of the membrane localization domain from Pasteurella multocida toxin | | Descriptor: | Dermonecrotic toxin | | Authors: | Hisao, G.S, Brothers, M.C, Ho, M, Wilson, B.A, Rienstra, C.M. | | Deposit date: | 2015-12-12 | | Release date: | 2016-12-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The membrane localization domains of two distinct bacterial toxins form a 4-helix-bundle in solution.
Protein Sci., 26, 2017
|
|
4G56
 
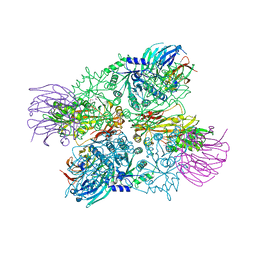 | | Crystal Structure of full length PRMT5/MEP50 complexes from Xenopus laevis | | Descriptor: | Hsl7 protein, MGC81050 protein, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Ho, M, Wilczek, C, Bonanno, J, Shechter, D, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-07-17 | | Release date: | 2012-10-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of the arginine methyltransferase PRMT5-MEP50 reveals a mechanism for substrate specificity
Plos One, 8, 2013
|
|
7CN9
 
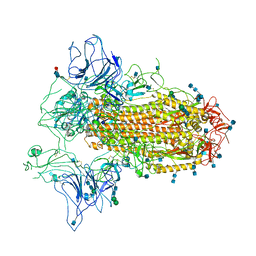 | | Cryo-EM structure of SARS-CoV-2 Spike ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ho, M, Chang, Y, Wang, C, Wu, Y, Huang, H, Chen, T, Lo, J.M, Chen, X, Ma, C. | | Deposit date: | 2020-07-30 | | Release date: | 2020-08-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | A Carbohydrate-Binding Protein from the Edible Lablab Beans Effectively Blocks the Infections of Influenza Viruses and SARS-CoV-2.
Cell Rep, 32, 2020
|
|
4GBD
 
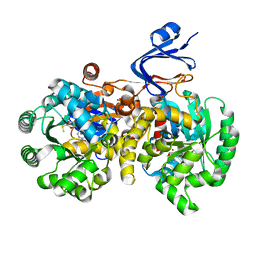 | | Crystal Structure Of Adenosine Deaminase From Pseudomonas Aeruginosa Pao1 with bound Zn and methylthio-coformycin | | Descriptor: | (8R)-3-(5-S-methyl-5-thio-beta-D-ribofuranosyl)-3,6,7,8-tetrahydroimidazo[4,5-d][1,3]diazepin-8-ol, PHOSPHATE ION, Putative uncharacterized protein, ... | | Authors: | Ho, M, Guan, R, Almo, S.C, Schramm, V.L. | | Deposit date: | 2012-07-27 | | Release date: | 2013-06-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.975 Å) | | Cite: | Methylthioadenosine deaminase in an alternative quorum sensing pathway in Pseudomonas aeruginosa.
Biochemistry, 51, 2012
|
|
3MB8
 
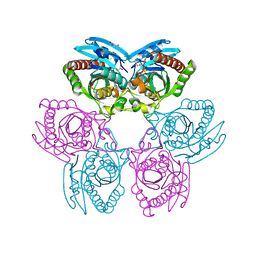 | | Crystal structure of purine nucleoside phosphorylase from toxoplasma gondii in complex with immucillin-H | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Ho, M, Almo, S.C, Schramm, V.L. | | Deposit date: | 2010-03-25 | | Release date: | 2011-04-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibition and Structure of Toxoplasma gondii Purine Nucleoside Phosphorylase.
Eukaryot Cell, 13, 2014
|
|
3OZF
 
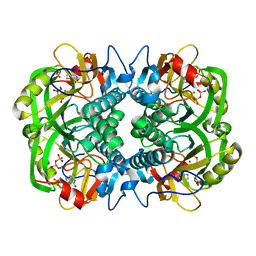 | | Crystal Structure of Plasmodium falciparum Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferase in complex with hypoxanthine | | Descriptor: | HYPOXANTHINE, Hypoxanthine-guanine-xanthine phosphoribosyltransferase, MAGNESIUM ION, ... | | Authors: | Ho, M, Hazleton, K.Z, Almo, S.C, Schramm, V.L. | | Deposit date: | 2010-09-24 | | Release date: | 2011-09-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | Acyclic Immucillin Phosphonates: Second-Generation Inhibitors of Plasmodium falciparum Hypoxanthine- Guanine-Xanthine Phosphoribosyltransferase.
Chem.Biol., 19, 2012
|
|
3OZB
 
 | | Crystal Structure of 5'-methylthioinosine phosphorylase from Psedomonas aeruginosa in complex with hypoxanthine | | Descriptor: | HYPOXANTHINE, Methylthioadenosine phosphorylase, SULFATE ION | | Authors: | Ho, M, Guan, R, Almo, S.C, Schramm, V.L. | | Deposit date: | 2010-09-24 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Methylthioinosine phosphorylase from Pseudomonas aeruginosa. Structure and annotation of a novel enzyme in quorum sensing.
Biochemistry, 50, 2011
|
|
3OZG
 
 | | Crystal Structure of Plasmodium falciparum Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferase in complex with S-SerMe-ImmH phosphonate | | Descriptor: | Hypoxanthine-guanine-xanthine phosphoribosyltransferase, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Ho, M, Hazleton, K.Z, Almo, S.C, Schramm, V.L. | | Deposit date: | 2010-09-24 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.993 Å) | | Cite: | Acyclic Immucillin Phosphonates: Second-Generation Inhibitors of Plasmodium falciparum Hypoxanthine- Guanine-Xanthine Phosphoribosyltransferase.
Chem.Biol., 19, 2012
|
|
3BH3
 
 | |
