8DOK
 
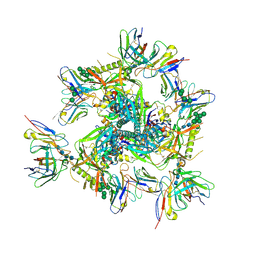 | | Cryo-EM structure of T/F100 SOSIP.664 HIV-1 Env trimer in complex with 8ANC195 and 10-1074 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, Y, Zhou, F, Huang, R, Tolbert, W, Pazgier, M. | | Deposit date: | 2022-07-13 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure-function analyses reveal key molecular determinants of HIV-1 CRF01_AE resistance to the entry inhibitor temsavir.
Nat Commun, 14, 2023
|
|
8G6U
 
 | | Cryo-EM structure of T/F100 SOSIP.664 HIV-1 Env trimer with LMHS mutations in complex with 8ANC195 and 10-1074 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CRF-1_AE T/F100 HIV-1 gp41, ... | | Authors: | Chen, Y, Zhou, F, Huang, R, Tolbert, W, Pazgier, M. | | Deposit date: | 2023-02-16 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structure-function analyses reveal key molecular determinants of HIV-1 CRF01_AE resistance to the entry inhibitor temsavir.
Nat Commun, 14, 2023
|
|
8TTW
 
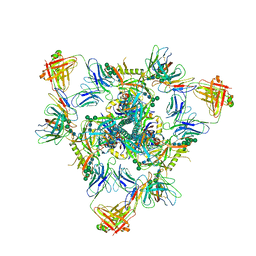 | | Cryo-EM structure of BG505 SOSIP.664 HIV-1 Env trimer in complex with temsavir, 8ANC195, and 10-1074 | | Descriptor: | 1-[4-(benzenecarbonyl)piperazin-1-yl]-2-[4-methoxy-7-(3-methyl-1H-1,2,4-triazol-1-yl)-1H-pyrrolo[2,3-c]pyridin-3-yl]ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tolbert, W.D, Pozharski, E, Pazgier, M. | | Deposit date: | 2023-08-15 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structure-function analyses reveal key molecular determinants of HIV-1 CRF01_AE resistance to the entry inhibitor temsavir.
Nat Commun, 14, 2023
|
|
8CZZ
 
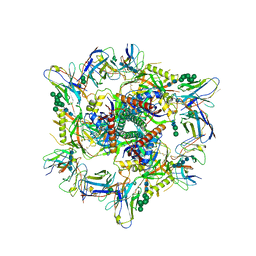 | | Cryo-EM structure of T/F100 SOSIP.664 HIV-1 Env trimer with LMHS mutations in complex with Temsavir, 8ANC195, and 10-1074 | | Descriptor: | 1-[4-(benzenecarbonyl)piperazin-1-yl]-2-[4-methoxy-7-(3-methyl-1H-1,2,4-triazol-1-yl)-1H-pyrrolo[2,3-c]pyridin-3-yl]ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, Y, Pozharski, E, Tolbert, W, Pazgier, M. | | Deposit date: | 2022-05-25 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structure-function analyses reveal key molecular determinants of HIV-1 CRF01_AE resistance to the entry inhibitor temsavir.
Nat Commun, 14, 2023
|
|
7UL1
 
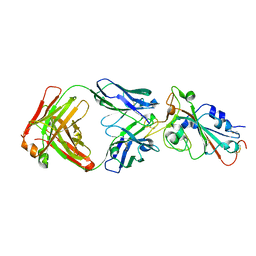 | | Crystal structure of SARS-CoV-2 RBD in complex with the neutralizing IGHV3-53-encoded antibody EH3 isolated from a nonvaccinated pediatric patient | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of EH3, Light chain of EH3, ... | | Authors: | Chen, Y, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2022-04-03 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Molecular basis for antiviral activity of two pediatric neutralizing antibodies targeting SARS-CoV-2 Spike RBD.
Iscience, 26, 2023
|
|
7UL0
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with the ridge-binding nAb EH8 isolated from a nonvaccinated pediatric patient | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of EH8, ... | | Authors: | Chen, Y, Tolbert, W.D, Bai, X, Pazgier, M. | | Deposit date: | 2022-04-03 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Molecular basis for antiviral activity of two pediatric neutralizing antibodies targeting SARS-CoV-2 Spike RBD.
Iscience, 26, 2023
|
|
3S6V
 
 | | Manganese-bound Ac-ASP-7 | | Descriptor: | Ac-ASP-7, MANGANESE (II) ION | | Authors: | Osman, A, Wang, C.K, Winter, A, Loukas, A, Tribolet, L, Gasser, R, Hofmann, A. | | Deposit date: | 2011-05-26 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Hookworm SCP/TAPS protein structure--A key to understanding host-parasite interactions and developing new interventions.
Biotechnol Adv, 30
|
|
3S6U
 
 | | Caclcium-bound Ac-ASP-7 | | Descriptor: | Ac-ASP-7, CALCIUM ION | | Authors: | Osman, A, Wang, C.K, Winter, A, Loukas, A, Tribolet, L, Gasser, R, Hofmann, A. | | Deposit date: | 2011-05-26 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Hookworm SCP/TAPS protein structure--A key to understanding host-parasite interactions and developing new interventions.
Biotechnol Adv, 30
|
|
3S6S
 
 | | Ancylostoma-secreted protein Ac-ASP-7 | | Descriptor: | Ac-ASP-7 | | Authors: | Osman, A, Wang, C.K, Winter, A, Loukas, A, Tribolet, L, Gasser, R, Hofmann, A. | | Deposit date: | 2011-05-26 | | Release date: | 2012-04-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hookworm SCP/TAPS protein structure--A key to understanding host-parasite interactions and developing new interventions.
Biotechnol Adv, 30
|
|
7NAB
 
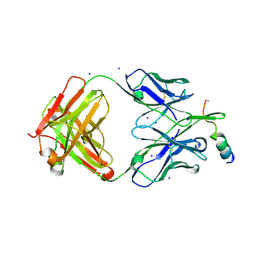 | | Crystal structure of human neutralizing mAb CV3-25 binding to SARS-CoV-2 S MPER peptide 1140-1165 | | Descriptor: | CITRIC ACID, CV3-25 Fab Heavy Chain, CV3-25 Fab Light Chain, ... | | Authors: | Chen, Y, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2021-06-21 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis and mode of action for two broadly neutralizing antibodies against SARS-CoV-2 emerging variants of concern.
Cell Rep, 38, 2022
|
|
5HZ5
 
 | |
5HZ6
 
 | |
5HZ8
 
 | | FABP4_3 in complex with 6,8-Dichloro-4-phenyl-2-piperidin-1-yl-quinoline-3-carboxylic acid | | Descriptor: | 6,8-dichloro-4-phenyl-2-(piperidin-1-yl)quinoline-3-carboxylic acid, DIMETHYL SULFOXIDE, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Rudolph, M.G. | | Deposit date: | 2016-02-02 | | Release date: | 2016-12-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Design and synthesis of selective, dual fatty acid binding protein 4 and 5 inhibitors.
Bioorg. Med. Chem. Lett., 26, 2016
|
|
5HZ9
 
 | |
6FER
 
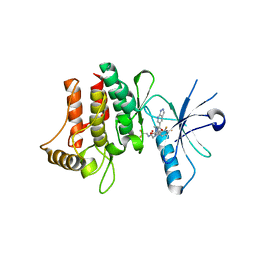 | | Crystal Structure of human DDR2 kinase in complex with 2-[4,5-difluoro-2-oxo-1'-(1H-pyrazolo[3,4-b]pyridine-5-carbonyl)spiro[indole-3,4'-piperidine]-1-yl]-N-(2,2,2-trifluoroethyl)acetamide | | Descriptor: | 2-[4,5-bis(fluoranyl)-2-oxidanylidene-1'-(1~{H}-pyrazolo[3,4-b]pyridin-5-ylcarbonyl)spiro[indole-3,4'-piperidine]-1-yl]-~{N}-[2,2,2-tris(fluoranyl)ethyl]ethanamide, Discoidin domain-containing receptor 2 | | Authors: | Stihle, M, Richter, H, Benz, J, Kuhn, B, Rudolph, M.G. | | Deposit date: | 2018-01-03 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | DNA-Encoded Library-Derived DDR1 Inhibitor Prevents Fibrosis and Renal Function Loss in a Genetic Mouse Model of Alport Syndrome.
Acs Chem.Biol., 14, 2019
|
|
6FEW
 
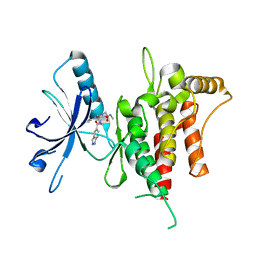 | | DDR1, 2-[8-(1H-indazole-5-carbonyl)-4-oxo-1-phenyl-1,3,8-triazaspiro[4.5]decan-3-yl]-N-methylacetamide, 1.440A, P1211, Rfree=24.1% | | Descriptor: | 2-[8-(2~{H}-indazol-5-ylcarbonyl)-4-oxidanylidene-1-phenyl-1,3,8-triazaspiro[4.5]decan-3-yl]-~{N}-methyl-ethanamide, Epithelial discoidin domain-containing receptor 1 | | Authors: | Stihle, M, Richter, H, Benz, J, Kuhn, B, Rudolph, M.G. | | Deposit date: | 2018-01-03 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | DNA-Encoded Library-Derived DDR1 Inhibitor Prevents Fibrosis and Renal Function Loss in a Genetic Mouse Model of Alport Syndrome.
Acs Chem.Biol., 14, 2019
|
|
6FIL
 
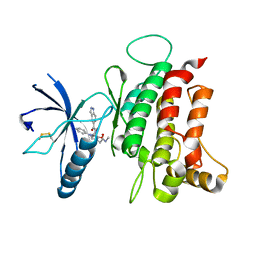 | | DDR1, 2-[8-(1H-indazole-5-carbonyl)-4-oxo-1-phenyl-1,3,8-triazaspiro[4.5]decan-3-yl]-N-methylacetamide, 1.730A, P212121, Rfree=24.5% | | Descriptor: | 2-[8-(2~{H}-indazol-5-ylcarbonyl)-4-oxidanylidene-1-phenyl-1,3,8-triazaspiro[4.5]decan-3-yl]-~{N}-methyl-ethanamide, Epithelial discoidin domain-containing receptor 1, IODIDE ION | | Authors: | Stihle, M, Richter, H, Benz, J, Kuhn, B, Rudolph, M.G. | | Deposit date: | 2018-01-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | DNA-Encoded Library-Derived DDR1 Inhibitor Prevents Fibrosis and Renal Function Loss in a Genetic Mouse Model of Alport Syndrome.
Acs Chem.Biol., 14, 2019
|
|
6FEX
 
 | | DDR1, 2-[4-bromo-2-oxo-1'-(1H-pyrazolo[4,3-b]pyridine-5-carbonyl)spiro[indole-3,4'-piperidine]-1-yl]-N-(2,2,2-trifluoroethyl)acetamide, 1.291A, P212121, Rfree=17.4% | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[4-bromanyl-2-oxidanylidene-1'-(1~{H}-pyrazolo[4,3-b]pyridin-5-ylcarbonyl)spiro[indole-3,4'-piperidine]-1-yl]-~{N}-[2,2,2-tris(fluoranyl)ethyl]ethanamide, CHLORIDE ION, ... | | Authors: | Stihle, M, Richter, H, Benz, J, Kuhn, B, Rudolph, M.G. | | Deposit date: | 2018-01-03 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.291 Å) | | Cite: | DNA-Encoded Library-Derived DDR1 Inhibitor Prevents Fibrosis and Renal Function Loss in a Genetic Mouse Model of Alport Syndrome.
Acs Chem.Biol., 14, 2019
|
|
6FIN
 
 | | DDR1, 3-[(3-cyclopropyl-1,2,4-oxadiazol-5-yl)methyl]-8-(1H-indazole-5-carbonyl)-1-phenyl-1,3,8-triazaspiro[4.5]decan-4-one, 1.670A, P1211, Rfree=22.8% | | Descriptor: | 3-[(3-cyclopropyl-1,2,4-oxadiazol-5-yl)methyl]-8-(1~{H}-indazol-5-ylcarbonyl)-1-phenyl-1,3,8-triazaspiro[4.5]decan-4-one, Epithelial discoidin domain-containing receptor 1, IODIDE ION | | Authors: | Stihle, M, Richter, H, Benz, J, Kuhn, B, Rudolph, M.G. | | Deposit date: | 2018-01-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | DNA-Encoded Library-Derived DDR1 Inhibitor Prevents Fibrosis and Renal Function Loss in a Genetic Mouse Model of Alport Syndrome.
Acs Chem.Biol., 14, 2019
|
|
6FIQ
 
 | | DDR1, 1-(1H-indazole-5-carbonyl)-5'-methoxy-1'-[2-oxo-2-[(2S)-2-(trifluoromethyl)pyrrolidin-1-yl]ethyl]spiro[piperidine-4,3'-pyrrolo[3,2-b]pyridine]-2'-one, 1.790A, P212121, Rfree=23.8% | | Descriptor: | 1-(1~{H}-indazol-5-ylcarbonyl)-5'-methoxy-1'-[2-oxidanylidene-2-[(2~{S})-2-(trifluoromethyl)pyrrolidin-1-yl]ethyl]spiro[piperidine-4,3'-pyrrolo[3,2-b]pyridine]-2'-one, CHLORIDE ION, Epithelial discoidin domain-containing receptor 1 | | Authors: | Stihle, M, Richter, H, Benz, J, Kuhn, B, Rudolph, M.G. | | Deposit date: | 2018-01-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | DNA-Encoded Library-Derived DDR1 Inhibitor Prevents Fibrosis and Renal Function Loss in a Genetic Mouse Model of Alport Syndrome.
Acs Chem.Biol., 14, 2019
|
|
6FIO
 
 | | DDR1, 2-[1'-(1H-indazole-5-carbonyl)-4-methyl-2-oxospiro[indole-3,4'-piperidine]-1-yl]-N-(2,2,2-trifluoroethyl)acetamide, 1.990A, P6522, Rfree=27.7% | | Descriptor: | 2-[1'-(1~{H}-indazol-5-ylcarbonyl)-4-methyl-2-oxidanylidene-spiro[indole-3,4'-piperidine]-1-yl]-~{N}-[2,2,2-tris(fluoranyl)ethyl]ethanamide, Epithelial discoidin domain-containing receptor 1 | | Authors: | Stihle, M, Richter, H, Benz, J, Kuhn, B, Rudolph, M.G. | | Deposit date: | 2018-01-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | DNA-Encoded Library-Derived DDR1 Inhibitor Prevents Fibrosis and Renal Function Loss in a Genetic Mouse Model of Alport Syndrome.
Acs Chem.Biol., 14, 2019
|
|
7RQ6
 
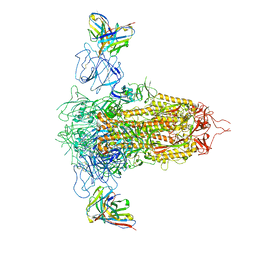 | | Cryo-EM structure of SARS-CoV-2 spike in complex with non-neutralizing NTD-directed CV3-13 Fab isolated from convalescent individual | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CV3-13 Fab heavy chain, ... | | Authors: | Chen, Y, Pozharski, E, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2021-08-05 | | Release date: | 2022-04-20 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | A Fc-enhanced NTD-binding non-neutralizing antibody delays virus spread and synergizes with a nAb to protect mice from lethal SARS-CoV-2 infection.
Cell Rep, 38, 2022
|
|
6T6I
 
 | |
6T6E
 
 | |
6T6J
 
 | |
