1IQ6
 
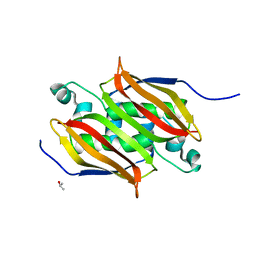 | | (R)-HYDRATASE FROM A. CAVIAE INVOLVED IN PHA BIOSYNTHESIS | | Descriptor: | (R)-SPECIFIC ENOYL-COA HYDRATASE, ISOPROPYL ALCOHOL | | Authors: | Hisano, T, Tsuge, T, Fukui, T, Iwata, T, Doi, Y. | | Deposit date: | 2001-06-18 | | Release date: | 2003-02-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the (R)-specific enoyl-CoA hydratase from Aeromonas caviae involved in polyhydroxyalkanoate
biosynthesis
J.Biol.Chem., 278, 2003
|
|
6LC9
 
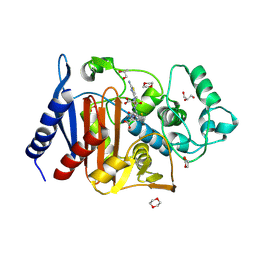 | | Crystal structure of AmpC Ent385 complex form with ceftazidime | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ACYLATED CEFTAZIDIME, Beta-lactamase, ... | | Authors: | Kawai, A, Doi, Y. | | Deposit date: | 2019-11-18 | | Release date: | 2020-04-22 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis of Reduced Susceptibility to Ceftazidime-Avibactam and Cefiderocol inEnterobacter cloacaeDue to AmpC R2 Loop Deletion.
Antimicrob.Agents Chemother., 64, 2020
|
|
6LC7
 
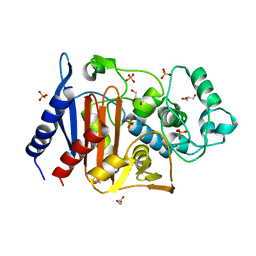 | | Crystal structure of AmpC Ent385 free form | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Beta-lactamase, GLYCEROL, ... | | Authors: | Kawai, A, Doi, Y. | | Deposit date: | 2019-11-18 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis of Reduced Susceptibility to Ceftazidime-Avibactam and Cefiderocol inEnterobacter cloacaeDue to AmpC R2 Loop Deletion.
Antimicrob.Agents Chemother., 64, 2020
|
|
1VMO
 
 | | CRYSTAL STRUCTURE OF VITELLINE MEMBRANE OUTER LAYER PROTEIN I (VMO-I): A FOLDING MOTIF WITH HOMOLOGOUS GREEK KEY STRUCTURES RELATED BY AN INTERNAL THREE-FOLD SYMMETRY | | Descriptor: | VITELLINE MEMBRANE OUTER LAYER PROTEIN I | | Authors: | Shimizu, T, Vassylyev, D.G, Kido, S, Doi, Y, Morikawa, K. | | Deposit date: | 1994-01-06 | | Release date: | 1994-05-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of vitelline membrane outer layer protein I (VMO-I): a folding motif with homologous Greek key structures related by an internal three-fold symmetry.
EMBO J., 13, 1994
|
|
8JB7
 
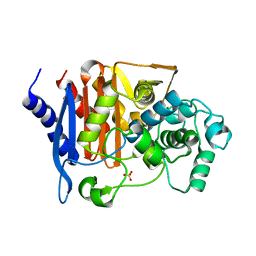 | | Crystal structure of CMY-185 | | Descriptor: | Beta-lactamase, SULFATE ION | | Authors: | Kawai, A, Doi, Y. | | Deposit date: | 2023-05-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural insights into the molecular mechanism of high-level ceftazidime-avibactam resistance conferred by CMY-185.
Mbio, 15, 2024
|
|
8JB8
 
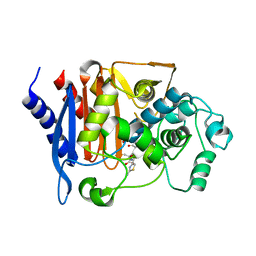 | | Crystal structure of CMY-185 complex with ceftazidime | | Descriptor: | ACYLATED CEFTAZIDIME, Beta-lactamase | | Authors: | Kawai, A, Doi, Y. | | Deposit date: | 2023-05-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the molecular mechanism of high-level ceftazidime-avibactam resistance conferred by CMY-185.
Mbio, 15, 2024
|
|
6LC8
 
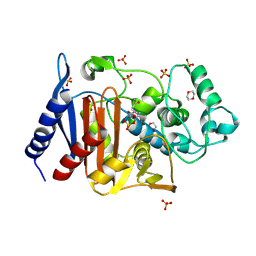 | | Crystal structure of AmpC Ent385 complex form with avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,4-DIETHYLENE DIOXIDE, Beta-lactamase, ... | | Authors: | Kawai, A, Doi, Y. | | Deposit date: | 2019-11-18 | | Release date: | 2020-04-22 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of Reduced Susceptibility to Ceftazidime-Avibactam and Cefiderocol inEnterobacter cloacaeDue to AmpC R2 Loop Deletion.
Antimicrob.Agents Chemother., 64, 2020
|
|
7EHF
 
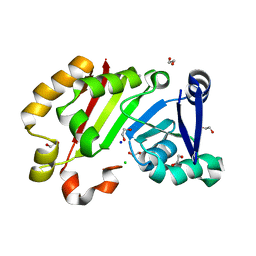 | | Crystal structure of the aminoglycoside resistance methyltransferase NpmB1 | | Descriptor: | 1,2-ETHANEDIOL, 16S rRNA methyltransferase, CHLORIDE ION, ... | | Authors: | Kawai, A, Doi, Y. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Functional and Structural Characterization of Acquired 16S rRNA Methyltransferase NpmB1 Conferring Pan-Aminoglycoside Resistance.
Antimicrob.Agents Chemother., 65, 2021
|
|
5WEP
 
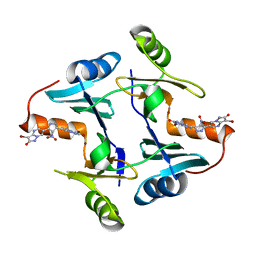 | | Crystal structure of fosfomycin resistance protein FosA3 with inhibitor (ANY1) bound | | Descriptor: | 6,6'-(4-nitro-1H-pyrazole-3,5-diyl)bis(3-bromopyrazolo[1,5-a]pyrimidin-2(1H)-one), FosA3, ZINC ION | | Authors: | Klontz, E.H, Sundberg, E.J. | | Deposit date: | 2017-07-10 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.502 Å) | | Cite: | Small-Molecule Inhibitor of FosA Expands Fosfomycin Activity to Multidrug-Resistant Gram-Negative Pathogens.
Antimicrob. Agents Chemother., 63, 2019
|
|
5WEW
 
 | | Crystal structure of Klebsiella pneumoniae fosfomycin resistance protein (FosAKP) with inhibitor (ANY1) bound | | Descriptor: | 6,6'-(4-nitro-1H-pyrazole-3,5-diyl)bis(3-bromopyrazolo[1,5-a]pyrimidin-2(1H)-one), Fosfomycin resistance protein, MANGANESE (II) ION | | Authors: | Klontz, E.H, Sundberg, E.J. | | Deposit date: | 2017-07-10 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.178 Å) | | Cite: | Small-Molecule Inhibitor of FosA Expands Fosfomycin Activity to Multidrug-Resistant Gram-Negative Pathogens.
Antimicrob. Agents Chemother., 63, 2019
|
|
8YXB
 
 | |
8YXA
 
 | | Crystal structure of the HSA complex with cefazolin and myristate | | Descriptor: | (6~{R},7~{R})-3-[(5-methyl-1,3,4-thiadiazol-2-yl)sulfanylmethyl]-8-oxidanylidene-7-[2-(1,2,3,4-tetrazol-1-yl)ethanoylamino]-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid, MYRISTIC ACID, Serum albumin | | Authors: | Kawai, A. | | Deposit date: | 2024-04-02 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Interaction of Cephalosporins with Human Serum Albumin: A Structural Study.
J.Med.Chem., 67, 2024
|
|
2ZJ9
 
 | | X-ray crystal structure of AmpC beta-Lactamase (AmpC(D)) from an Escherichia coli with a Tripeptide Deletion (Gly286 Ser287 Asp288) on the H10 Helix | | Descriptor: | AmpC, ISOPROPYL ALCOHOL, SODIUM ION | | Authors: | Yamaguchi, Y, Sato, G, Yamagata, Y, Wachino, J, Arakawa, Y, Kurosaki, H. | | Deposit date: | 2008-02-29 | | Release date: | 2009-03-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of AmpC beta-lactamase (AmpCD) from an Escherichia coli clinical isolate with a tripeptide deletion (Gly286-Ser287-Asp288) in the H10 helix
Acta Crystallogr.,Sect.F, 65, 2009
|
|
8JQO
 
 | | Protocatecuate hydroxylase from Xylophilus ampelinus complexed with imidazole | | Descriptor: | 4-hydroxybenzoate 3-monooxygenase (NAD(P)H), CALCIUM ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Fukushima, R, Katsuki, N, Fushinobu, S, Takaya, N. | | Deposit date: | 2023-06-14 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Protocatechuate hydroxylase is a novel group A flavoprotein monooxygenase with a unique substrate recognition mechanism.
J.Biol.Chem., 300, 2023
|
|
8JRD
 
 | | Chalcone synthase from Glycine max (L.) Merr (soybean) complexed with naringenin and coenzyme A | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Waki, T, Imaizumi, R, Nakata, S, Yanai, T, Takeshita, K, Sakai, N, Kataoka, K, Yamamoto, M, Nakayama, T, Yamashita, S. | | Deposit date: | 2023-06-16 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural insights into catalytic promiscuity of chalcone synthase from Glycine max (L.) Merr.: Coenzyme A-induced alteration of product specificity.
Biochem.Biophys.Res.Commun., 718, 2024
|
|
8JQQ
 
 | | Protocatecuate hydroxylase from Xylophilus ampelinus C347T mutant | | Descriptor: | 4-hydroxybenzoate 3-monooxygenase (NAD(P)H), CALCIUM ION, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fukushima, R, Katsuki, N, Fushinobu, S, Takaya, N. | | Deposit date: | 2023-06-14 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Protocatechuate hydroxylase is a novel group A flavoprotein monooxygenase with a unique substrate recognition mechanism.
J.Biol.Chem., 300, 2023
|
|
8JQP
 
 | | Protocatecuate hydroxylase from Xylophilus ampelinus complexed with 3,4-dihydroxybenzoate | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, 4-hydroxybenzoate 3-monooxygenase (NAD(P)H), CALCIUM ION, ... | | Authors: | Fukushima, R, Katsuki, N, Fushinobu, S, Takaya, N. | | Deposit date: | 2023-06-14 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Protocatechuate hydroxylase is a novel group A flavoprotein monooxygenase with a unique substrate recognition mechanism.
J.Biol.Chem., 300, 2023
|
|
5V91
 
 | | Crystal structure of fosfomycin resistance protein from Klebsiella pneumoniae | | Descriptor: | Fosfomycin resistance protein, ZINC ION | | Authors: | Klontz, E, Guenther, S, Silverstein, Z, Sundberg, E. | | Deposit date: | 2017-03-22 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure and Dynamics of FosA-Mediated Fosfomycin Resistance in Klebsiella pneumoniae and Escherichia coli.
Antimicrob. Agents Chemother., 61, 2017
|
|
5V3D
 
 | | Crystal structure of fosfomycin resistance protein from Klebsiella pneumoniae with bound fosfomycin | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FOSFOMYCIN, ... | | Authors: | Klontz, E, Guenther, S, Silverstein, Z, Sundberg, E. | | Deposit date: | 2017-03-07 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.539 Å) | | Cite: | Structure and Dynamics of FosA-Mediated Fosfomycin Resistance in Klebsiella pneumoniae and Escherichia coli.
Antimicrob. Agents Chemother., 61, 2017
|
|
5VB0
 
 | | Crystal structure of fosfomycin resistance protein FosA3 | | Descriptor: | Fosfomycin resistance protein FosA3, MANGANESE (II) ION, NICKEL (II) ION | | Authors: | Klontz, E, Guenther, S, Silverstein, Z, Sundberg, E. | | Deposit date: | 2017-03-28 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.689 Å) | | Cite: | Structure and Dynamics of FosA-Mediated Fosfomycin Resistance in Klebsiella pneumoniae and Escherichia coli.
Antimicrob. Agents Chemother., 61, 2017
|
|
6C3U
 
 | |
2D81
 
 | | PHB depolymerase (S39A) complexed with R3HB trimer | | Descriptor: | (1R)-3-{[(1R)-3-METHOXY-1-METHYL-3-OXOPROPYL]OXY}-1-METHYL-3-OXOPROPYL (3R)-3-HYDROXYBUTANOATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, PHB depolymerase | | Authors: | Hisano, T, Kasuya, K, Saito, T, Iwata, T, Miki, K. | | Deposit date: | 2005-11-30 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The Crystal Structure of Polyhydroxybutyrate Depolymerase from Penicillium funiculosum Provides Insights into the Recognition and Degradation of Biopolyesters
J.Mol.Biol., 356, 2006
|
|
2D7T
 
 | |
2D80
 
 | | Crystal structure of PHB depolymerase from Penicillium funiculosum | | Descriptor: | PHB depolymerase, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Hisano, T, Kasuya, K, Saito, T, Iwata, T, Miki, K. | | Deposit date: | 2005-11-30 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Polyhydroxybutyrate Depolymerase from Penicillium funiculosum Provides Insights into the Recognition and Degradation of Biopolyesters
J.Mol.Biol., 356, 2006
|
|
