7MN6
 
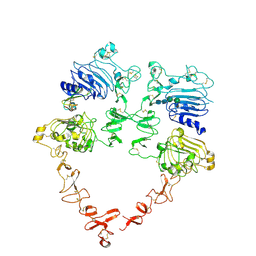 | | Structure of the HER2 S310F/HER3/NRG1b Heterodimer Extracellular Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 6 of Pro-neuregulin-1, ... | | Authors: | Diwanji, D, Trenker, R, Verba, K.A, Jura, N. | | Deposit date: | 2021-04-30 | | Release date: | 2021-10-27 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structures of the HER2-HER3-NRG1 beta complex reveal a dynamic dimer interface.
Nature, 600, 2021
|
|
7MN5
 
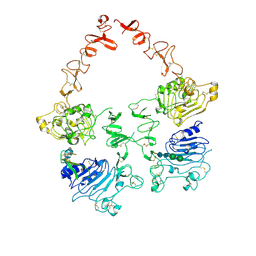 | | Structure of the HER2/HER3/NRG1b Heterodimer Extracellular Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 6 of Pro-neuregulin-1, ... | | Authors: | Diwanji, D, Trenker, R, Verba, K.A, Jura, N. | | Deposit date: | 2021-04-30 | | Release date: | 2021-10-27 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structures of the HER2-HER3-NRG1 beta complex reveal a dynamic dimer interface.
Nature, 600, 2021
|
|
7MN8
 
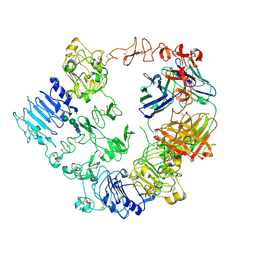 | | Structure of the HER2/HER3/NRG1b Heterodimer Extracellular Domain bound to Trastuzumab Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 6 of Pro-neuregulin-1, ... | | Authors: | Diwanji, D, Trenker, R, Verba, K.A, Jura, N. | | Deposit date: | 2021-04-30 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structures of the HER2-HER3-NRG1 beta complex reveal a dynamic dimer interface.
Nature, 600, 2021
|
|
5KZC
 
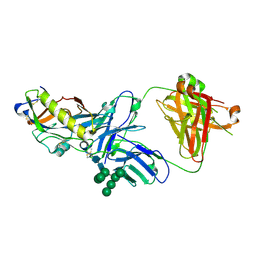 | | Crystal structure of an HIV-1 gp120 engineered outer domain with a Man9 glycan at position N276, in complex with broadly neutralizing antibody VRC01 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Engineered outer domain of gp120, VRC01 Fab heavy chain, ... | | Authors: | Julien, J.-P, Jardine, J.G, Diwanji, D, Schief, W.R, Wilson, I.A. | | Deposit date: | 2016-07-24 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Minimally Mutated HIV-1 Broadly Neutralizing Antibodies to Guide Reductionist Vaccine Design.
Plos Pathog., 12, 2016
|
|
8U4K
 
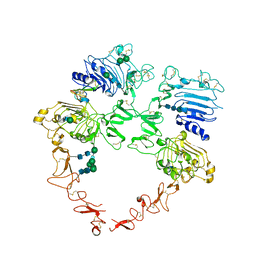 | | Structure of the HER2/HER4/BTC Heterodimer Extracellular Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Betacellulin, ... | | Authors: | Trenker, R, Diwanji, D, Bingham, T, Verba, K.A, Jura, N. | | Deposit date: | 2023-09-10 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Structural dynamics of the active HER4 and HER2/HER4 complexes is finely tuned by different growth factors and glycosylation.
Elife, 12, 2024
|
|
8U4I
 
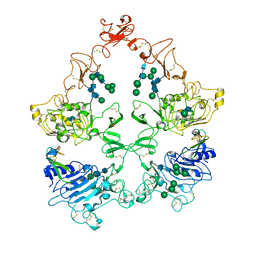 | | Structure of the HER4/NRG1b Homodimer Extracellular Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 6 of Pro-neuregulin-1, ... | | Authors: | Trenker, R, Diwanji, D, Bingham, T, Verba, K.A, Jura, N. | | Deposit date: | 2023-09-10 | | Release date: | 2024-03-13 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural dynamics of the active HER4 and HER2/HER4 complexes is finely tuned by different growth factors and glycosylation.
Elife, 12, 2024
|
|
8U4L
 
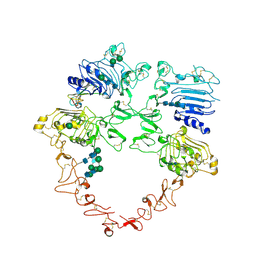 | | Structure of the HER2/HER4/NRG1b Heterodimer Extracellular Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 6 of Pro-neuregulin-1, ... | | Authors: | Trenker, R, Diwanji, D, Bingham, T, Verba, K.A, Jura, N. | | Deposit date: | 2023-09-10 | | Release date: | 2024-03-13 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structural dynamics of the active HER4 and HER2/HER4 complexes is finely tuned by different growth factors and glycosylation.
Elife, 12, 2024
|
|
8U4J
 
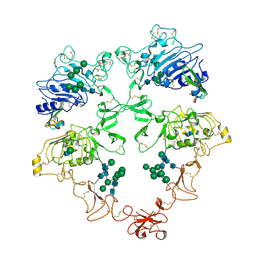 | | Structure of the HER4/BTC Homodimer Extracellular Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Betacellulin, ... | | Authors: | Trenker, R, Diwanji, D, Bingham, T, Verba, K.A, Jura, N. | | Deposit date: | 2023-09-10 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural dynamics of the active HER4 and HER2/HER4 complexes is finely tuned by different growth factors and glycosylation.
Elife, 12, 2024
|
|
7KKJ
 
 | | Structure of anti-SARS-CoV-2 Spike nanobody mNb6 | | Descriptor: | CHLORIDE ION, SULFATE ION, Synthetic nanobody mNb6 | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
7KKK
 
 | | SARS-CoV-2 Spike in complex with neutralizing nanobody Nb6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
7KKL
 
 | | SARS-CoV-2 Spike in complex with neutralizing nanobody mNb6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
3U1S
 
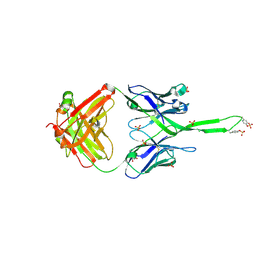 | | Crystal structure of human Fab PGT145, a broadly reactive and potent HIV-1 neutralizing antibody | | Descriptor: | Fab PGT145 Heavy chain, Fab PGT145 Light chain, GLYCEROL, ... | | Authors: | Julien, J.-P, Diwanji, D, Burton, D.R, Wilson, I.A. | | Deposit date: | 2011-09-30 | | Release date: | 2011-12-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
5D9Q
 
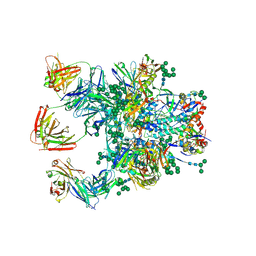 | | Crystal Structure of the BG505 SOSIP gp140 HIV-1 Env trimer in Complex with the Broadly Neutralizing Fab PGT122 and scFv NIH45-46 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Julien, J.-P, Stanfield, R.L, Ward, A.B, Wilson, I.A. | | Deposit date: | 2015-08-18 | | Release date: | 2016-08-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Minimally Mutated HIV-1 Broadly Neutralizing Antibodies to Guide Reductionist Vaccine Design.
Plos Pathog., 12, 2016
|
|
7KDT
 
 | |
8DP5
 
 | | Structure of the PEAK3/14-3-3 complex | | Descriptor: | 14-3-3 protein beta/alpha, 14-3-3 protein epsilon, Protein PEAK3, ... | | Authors: | Torosyan, H, Paul, M, Jura, N, Verba, K.A. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into regulation of the PEAK3 pseudokinase scaffold by 14-3-3.
Nat Commun, 14, 2023
|
|
8DS6
 
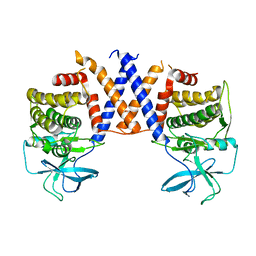 | |
8DV1
 
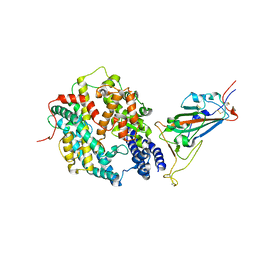 | | SARS-CoV-2 Wuhan-hu-1-Spike-RBD bound to linker variant of affinity matured ACE2 mimetic CVD432 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Immunoglobulin gamma-1 heavy chain fusion,Immunoglobulin gamma-1 heavy chain, Spike glycoprotein | | Authors: | QCRG Structural Biology Consortium, Remesh, S.G, Merz, G.E, Brilot, A.F, Chio, U, Verba, K.A. | | Deposit date: | 2022-07-27 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Computational pipeline provides mechanistic understanding of Omicron variant of concern neutralizing engineered ACE2 receptor traps.
Structure, 31, 2023
|
|
8DV2
 
 | | SARS-CoV-2 Wuhan-hu-1-Spike-RBD bound to computationally engineered ACE2 mimetic CVD293 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Immunoglobulin gamma-1 heavy chain fusion, Spike glycoprotein | | Authors: | QCRG Structural Biology Consortium, Remesh, S.G, Merz, G.E, Brilot, A.F, Chio, U, Verba, K.A. | | Deposit date: | 2022-07-27 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Computational pipeline provides mechanistic understanding of Omicron variant of concern neutralizing engineered ACE2 receptor traps.
Structure, 31, 2023
|
|
3U46
 
 | | CH04H/CH02L P212121 | | Descriptor: | CH02 Light chain Fab, CH04 Heavy chain Fab | | Authors: | Louder, R, Pancera, M, McLellan, J.S, Kwong, P.D. | | Deposit date: | 2011-10-07 | | Release date: | 2011-11-30 | | Last modified: | 2011-12-21 | | Method: | X-RAY DIFFRACTION (2.906 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
3TCL
 
 | | Crystal Structure of HIV-1 Neutralizing Antibody CH04 | | Descriptor: | CH04 Heavy Chain Fab, CH04 Light Chain Fab, IMIDAZOLE | | Authors: | Louder, R.K, McLellan, J.S, Pancera, M, Yang, Y, Zhang, B, Bonsignori, M, Kwong, P.D. | | Deposit date: | 2011-08-09 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
3U4E
 
 | | Crystal Structure of PG9 Fab in Complex with V1V2 Region from HIV-1 strain CAP45 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PG9 Heavy Chain, PG9 Light Chain, ... | | Authors: | Gorman, J, McLellan, J, Pancera, M, Kwong, P.D. | | Deposit date: | 2011-10-07 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.185 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
3U4B
 
 | | CH04H/CH02L Fab P4 | | Descriptor: | CH02 Light chain, CH04 Heavy chain | | Authors: | Pancera, M, Louder, R, Mclellan, J.S, KWong, P.D. | | Deposit date: | 2011-10-07 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.893 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
3U36
 
 | | Crystal Structure of PG9 Fab | | Descriptor: | PG9 Fab heavy chain, PG9 Fab light chain, SULFATE ION | | Authors: | McLellan, J.S, Kwong, P.D. | | Deposit date: | 2011-10-04 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.281 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
3U2S
 
 | | Crystal Structure of PG9 Fab in Complex with V1V2 Region from HIV-1 strain ZM109 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | McLellan, J.S, Pancera, M, Kwong, P.D. | | Deposit date: | 2011-10-04 | | Release date: | 2011-11-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
