8PAY
 
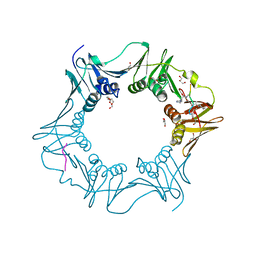 | | Structure of the E.coli DNA polymerase sliding clamp with a covalently bound peptide 2. | | Descriptor: | ACE-GLN-ALC-GLC-LEU-PHE, Beta sliding clamp, GLYCEROL, ... | | Authors: | Compain, G, Monsarrat, C, Blagojevic, J, Brillet, K, Dumas, P, Hammann, P, Kuhn, L, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Burnouf, D, wagner, J, Guichard, G. | | Deposit date: | 2023-06-08 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Peptide-Based Covalent Inhibitors Bearing Mild Electrophiles to Target a Conserved His Residue of the Bacterial Sliding Clamp.
Jacs Au, 4, 2024
|
|
8PAT
 
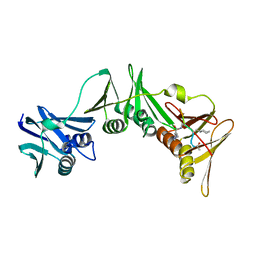 | | Structure of the E.coli DNA polymerase sliding clamp with a covalently bound peptide 3. | | Descriptor: | ACE-GLN-ALC-GLX-LEU-PHE, Beta sliding clamp | | Authors: | Compain, G, Monsarrat, C, Blagojevic, J, Brillet, K, Dumas, P, Hammann, P, Kuhn, L, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Burnouf, D, Guichard, G. | | Deposit date: | 2023-06-08 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Peptide-Based Covalent Inhibitors Bearing Mild Electrophiles to Target a Conserved His Residue of the Bacterial Sliding Clamp.
Jacs Au, 4, 2024
|
|
7AZ5
 
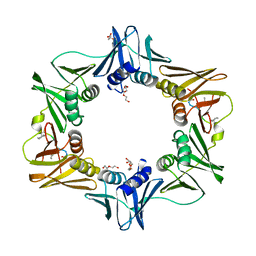 | | DNA polymerase sliding clamp from Escherichia coli with peptide 47 bound | | Descriptor: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, Peptide 47, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZG
 
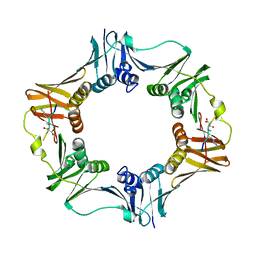 | | DNA polymerase sliding clamp from Escherichia coli with peptide 4 bound | | Descriptor: | Beta sliding clamp, Peptide 4 | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZL
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 38 bound | | Descriptor: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZ7
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 37 bound | | Descriptor: | Beta sliding clamp, FORMIC ACID, PENTAETHYLENE GLYCOL, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZE
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 18 bound | | Descriptor: | Beta sliding clamp, GLYCEROL, MALONATE ION, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZ6
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 36 bound | | Descriptor: | ACETATE ION, Beta sliding clamp, CHLORIDE ION, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZ8
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 43 bound | | Descriptor: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZC
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 22 bound | | Descriptor: | Beta sliding clamp, GLYCEROL, Peptide 22 | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZF
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 8 bound | | Descriptor: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZD
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 20 bound | | Descriptor: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZK
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 35 bound | | Descriptor: | Beta sliding clamp, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
4Y59
 
 | | Crystal structure of ALiS1-Streptavidin complex | | Descriptor: | 2-[3-(trifluoromethyl)phenyl]furo[3,2-c]pyridin-4(5H)-one, Streptavidin | | Authors: | Sugiyama, S, Terai, T, Kohno, M, Ishida, H, Nagano, T. | | Deposit date: | 2015-02-11 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Artificial Ligands of Streptavidin (ALiS): Discovery, Characterization, and Application for Reversible Control of Intracellular Protein Transport
J.Am.Chem.Soc., 137, 2015
|
|
4Y5D
 
 | | CRYSTAL STRUCTURE OF ALiS2-STREPTAVIDIN COMPLEX | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, DIMETHYL SULFOXIDE, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Terai, T, Kohno, M, Ishida, H, Nagano, T. | | Deposit date: | 2015-02-11 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Artificial Ligands of Streptavidin (ALiS): Discovery, Characterization, and Application for Reversible Control of Intracellular Protein Transport
J.Am.Chem.Soc., 137, 2015
|
|
5B5G
 
 | | Crystal structure of ALiS4-Streptavidin complex | | Descriptor: | SULFITE ION, Streptavidin, methyl 5-(4-oxidanylidene-5~{H}-furo[3,2-c]pyridin-2-yl)pyridine-3-carboxylate | | Authors: | Sugiyama, S, Terai, T, Kakinouchi, K, Fujikake, R, Nagano, T, Urano, Y. | | Deposit date: | 2016-05-04 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Improving the Solubility of Artificial Ligands of Streptavidin to Enable More Practical Reversible Switching of Protein Localization in Cells
Chembiochem, 18, 2017
|
|
5B5F
 
 | | Crystal structure of ALiS3-Streptavidin complex | | Descriptor: | N-methyl-3-(4-oxo-4,5-dihydrofuro[3,2-c]pyridin-2-yl)benzenesulfonamide, Streptavidin | | Authors: | Sugiyama, S, Terai, T, Kakinouchi, K, Fujikake, R, Nagano, T, Urano, Y. | | Deposit date: | 2016-05-04 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Improving the Solubility of Artificial Ligands of Streptavidin to Enable More Practical Reversible Switching of Protein Localization in Cells
Chembiochem, 18, 2017
|
|
