8B8G
 
 | | Cryo-EM structure of Ca2+-free mTMEM16F F518H mutant in Digitonin | | Descriptor: | Anoctamin-6 | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8B8K
 
 | | Cryo-EM structure of Ca2+-bound mTMEM16F N562A mutant in Digitonin closed/closed | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Anoctamin-6, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8BC0
 
 | | Cryo-EM structure of Ca2+-bound mTMEM16F F518A Q623A mutant in GDN open/closed | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-14 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8BC1
 
 | | Cryo-EM Structure of Ca2+-bound mTMEM16F F518A_Q623A mutant in GDN | | Descriptor: | Anoctamin-6,mTMEM16F, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-14 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8B8M
 
 | | Cryo-EM structure of Ca2+-bound mTMEM16F N562A mutant in Digitonin open/closed | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8B8Q
 
 | | Structure of mTMEM16F in lipid Nanodiscs in the presence of Ca2+ | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
6QMB
 
 | | Cryo-EM structure of calcium-bound nhTMEM16 lipid scramblase in nanodisc (closed state) | | Descriptor: | CALCIUM ION, Predicted protein | | Authors: | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-01 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
6QM9
 
 | | Cryo-EM structure of calcium-bound nhTMEM16 lipid scramblase in nanodisc (open state) | | Descriptor: | CALCIUM ION, Predicted protein | | Authors: | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-01 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
6QM4
 
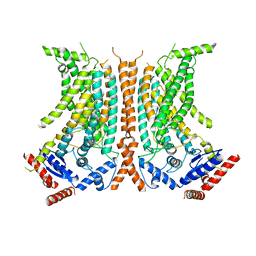 | | Cryo-EM structure of calcium-free nhTMEM16 lipid scramblase in nanodisc | | Descriptor: | Predicted protein | | Authors: | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-01 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
6QM6
 
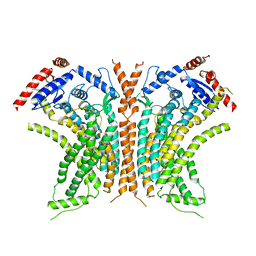 | | Cryo-EM structure of calcium-free nhTMEM16 lipid scramblase in DDM | | Descriptor: | Predicted protein | | Authors: | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-01 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
6QMA
 
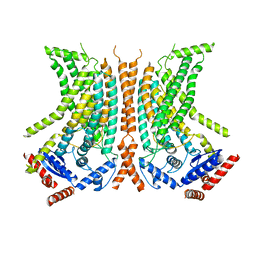 | | Cryo-EM structure of calcium-bound nhTMEM16 lipid scramblase in nanodisc (intermediate state) | | Descriptor: | CALCIUM ION, Predicted protein | | Authors: | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-01 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
6QM5
 
 | | Cryo-EM structure of calcium-bound nhTMEM16 lipid scramblase in DDM | | Descriptor: | CALCIUM ION, Predicted protein | | Authors: | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-01 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
6QPB
 
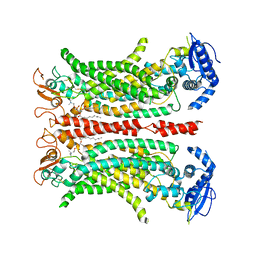 | | Cryo-EM structure of calcium-free mTMEM16F lipid scramblase in digitonin | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Anoctamin-6 | | Authors: | Alvadia, C, Lim, N.K, Clerico Mosina, V, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-13 | | Release date: | 2019-03-06 | | Last modified: | 2019-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures and functional characterization of the murine lipid scramblase TMEM16F.
Elife, 8, 2019
|
|
6QP6
 
 | | Cryo-EM structure of calcium-bound mTMEM16F lipid scramblase in digitonin | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Anoctamin-6, CALCIUM ION | | Authors: | Alvadia, C, Lim, N.K, Clerico Mosina, V, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-13 | | Release date: | 2019-03-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures and functional characterization of the murine lipid scramblase TMEM16F.
Elife, 8, 2019
|
|
6QPI
 
 | | Cryo-EM structure of calcium-free mTMEM16F lipid scramblase in nanodisc | | Descriptor: | Anoctamin-6 | | Authors: | Alvadia, C, Lim, N.K, Clerico Mosina, V, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-14 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures and functional characterization of the murine lipid scramblase TMEM16F.
Elife, 8, 2019
|
|
6QPC
 
 | | Cryo-EM structure of calcium-bound mTMEM16F lipid scramblase in nanodisc | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Anoctamin-6, CALCIUM ION | | Authors: | Alvadia, C, Lim, N.K, Clerico Mosina, V, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-13 | | Release date: | 2019-03-06 | | Last modified: | 2019-04-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures and functional characterization of the murine lipid scramblase TMEM16F.
Elife, 8, 2019
|
|
8B8J
 
 | | Cryo-EM structure of Ca2+-bound mTMEM16F F518H mutant in Digitonin | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
