4USC
 
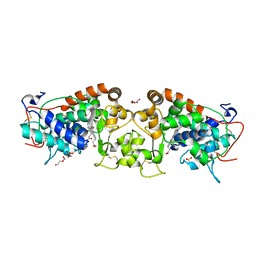 | | Crystal structure of peroxidase from palm tree Chamaerops excelsa | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Bernardes, A, Santos, J.C, Textor, L.C, Cuadrado, N.H, Kostetsky, E.Y, Roig, M.G, Muniz, J.R.C, Shnyrov, V.L, Polikarpov, I. | | Deposit date: | 2014-07-07 | | Release date: | 2015-05-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure Analysis of Peroxidase from the Palm Tree Chamaerops Excelsa.
Biochimie, 111, 2015
|
|
4BCR
 
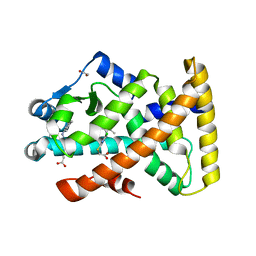 | | Structure of PPARalpha in complex with WY14643 | | Descriptor: | 1,2-ETHANEDIOL, 2-({4-CHLORO-6-[(2,3-DIMETHYLPHENYL)AMINO]PYRIMIDIN-2-YL}SULFANYL)ACETIC ACID, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR ALPHA | | Authors: | Bernardes, A, Muniz, J.R.C, Polikarpov, I. | | Deposit date: | 2012-10-02 | | Release date: | 2013-05-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Molecular Mechanism of Peroxisome Proliferator-Activated Receptor Alpha Activation by Wy14643: A New Mode of Ligand Recognition and Receptor Stabilization
J.Mol.Biol., 425, 2013
|
|
2V3Q
 
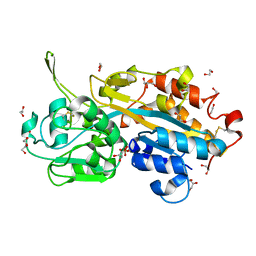 | | Serendipitous discovery and X-ray structure of a human phosphate binding apolipoprotein | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, HUMAN PHOSPHATE BINDING PROTEIN, ... | | Authors: | Morales, R, Berna, A, Carpentier, P, Elias, M, Contreras-Martel, C, Renault, F, Nicodeme, M, Chesne-Seck, M.-L, Bernier, F, Dupuy, J, Schaeffer, C, Diemer, H, Van Dorsselaer, A, Fontecilla, J.C, Masson, P, Rochu, D, Chabriere, E. | | Deposit date: | 2007-06-20 | | Release date: | 2008-07-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Tandem Use of X-Ray Crystallography and Mass Spectrometry to Obtain Ab Initio the Complete and Exact Amino Acids Sequence of Hpbp, a Human 38kDa Apolipoprotein
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
6R0N
 
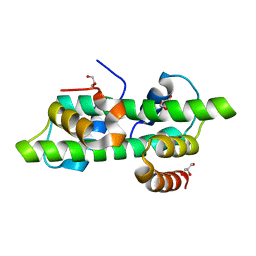 | | Histone fold domain of AtNF-YB2/NF-YC3 in I2 | | Descriptor: | GLYCEROL, NF-YB2, NF-YC3 | | Authors: | Chaves-Sanjuan, A, Gnesutta, N, Bernardini, A, Fornara, F, Nardini, M, Mantovani, R. | | Deposit date: | 2019-03-13 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural determinants for NF-Y subunit organization and NF-Y/DNA association in plants.
Plant J., 105, 2021
|
|
6R0M
 
 | | Histone fold domain of AtNF-YB2/NF-YC3 in P212121 | | Descriptor: | NF-YB2, NF-YC3 | | Authors: | Chaves-Sanjuan, A, Gnesutta, N, Chiara, M, Bernardini, A, Fornara, F, Horner, D, Nardini, M, Mantovani, R. | | Deposit date: | 2019-03-13 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural determinants for NF-Y subunit organization and NF-Y/DNA association in plants.
Plant J., 105, 2021
|
|
6R2V
 
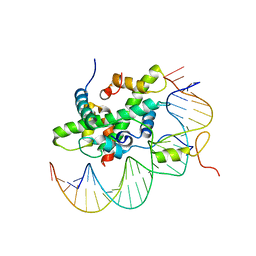 | | Arabidopsis NF-Y/CCAAT-box complex | | Descriptor: | FT (-5kb) CCAAT-box 3', FT (-5kb) CCAAT-box 5', NF-YB2, ... | | Authors: | Chaves-Sanjuan, A, Gnesutta, N, Chiara, M, Bernardini, A, Fornara, F, Horner, D, Nardini, M, Mantovani, R. | | Deposit date: | 2019-03-19 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Structural determinants for NF-Y subunit organization and NF-Y/DNA association in plants.
Plant J., 105, 2021
|
|
8RCY
 
 | | L-SIGN CRD in complex with Man84. | | Descriptor: | 1-[[1-[(2S,3S,4R,5S,6R)-2-(2-chloroethyloxy)-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-3-yl]-1,2,3-triazol-4-yl]methyl]guanidine, C-type lectin domain family 4 member M, CALCIUM ION, ... | | Authors: | Thepaut, M, Bouchikri, C, Pollastri, S, Bernardi, A, Fieschi, F. | | Deposit date: | 2023-12-07 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unprecedented selectivity for homologous lectin targets: differential targeting of the viral receptors L-SIGN and DC-SIGN.
Chem Sci, 15, 2024
|
|
8QO4
 
 | | Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes | | Descriptor: | MERS-CoV-SL5 | | Authors: | Moura, T.R, Purta, E, Bernat, A, Baulin, E, Mukherjee, S, Bujnicki, J.M. | | Deposit date: | 2023-09-28 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Conserved structures and dynamics in 5'-proximal regions of Betacoronavirus RNA genomes.
Nucleic Acids Res., 52, 2024
|
|
8QO2
 
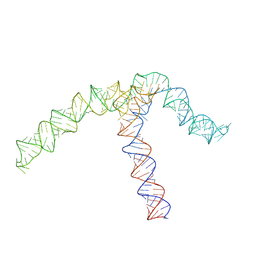 | | Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes | | Descriptor: | OC43-CoV-SL5 | | Authors: | Moura, T.R, Purta, E, Bernat, A, Baulin, E, Mukherjee, S, Bujnicki, J.M. | | Deposit date: | 2023-09-27 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Conserved structures and dynamics in 5'-proximal regions of Betacoronavirus RNA genomes.
Nucleic Acids Res., 52, 2024
|
|
8QO3
 
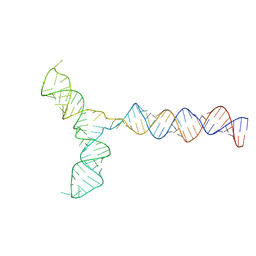 | | Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes | | Descriptor: | RoBat-CoV-SL5 | | Authors: | Moura, T.R, Purta, E, Bernat, A, Baulin, E, Mukherjee, S, Bujnicki, J.M. | | Deposit date: | 2023-09-28 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Conserved structures and dynamics in 5'-proximal regions of Betacoronavirus RNA genomes.
Nucleic Acids Res., 52, 2024
|
|
8QO5
 
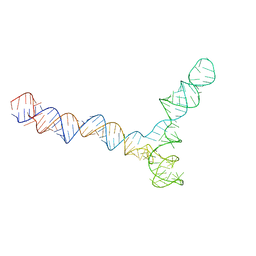 | | Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes | | Descriptor: | SARS-CoV-2-SL5 | | Authors: | Moura, T.R, Purta, E, Bernat, A, Baulin, E, Mukherjee, S, Bujnicki, J.M. | | Deposit date: | 2023-09-28 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Conserved structures and dynamics in 5'-proximal regions of Betacoronavirus RNA genomes.
Nucleic Acids Res., 52, 2024
|
|
4WX4
 
 | | Crystal structure of adenovirus 8 protease in complex with a nitrile inhibitor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCINE, N-[(2-cyanopyrimidin-4-yl)methyl]-3-[2-(3,5-dichlorophenyl)-2-methylpropanoyl]-4-methoxybenzamide, ... | | Authors: | Grosche, P, Sirockin, F, Mac Sweeney, A, Ramage, P, Erbel, P, Melkko, S, Bernardi, A, Hughes, N, Ellis, D, Combrink, K, Jarousse, N, Altmann, E. | | Deposit date: | 2014-11-13 | | Release date: | 2015-01-14 | | Last modified: | 2015-01-28 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Structure-based design and optimization of potent inhibitors of the adenoviral protease.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4WX6
 
 | | Crystal structure of human adenovirus 8 protease with an irreversible vinyl sulfone inhibitor | | Descriptor: | N-[(2S)-2-(3,5-dichlorophenyl)-2-(ethylamino)acetyl]-3-methyl-L-valyl-N-[3-(methylsulfonyl)propyl]glycinamide, PVI, Protease | | Authors: | Grosche, P, Sirockin, F, Mac Sweeney, A, Ramage, P, Erbel, P, Melkko, S, Bernardi, A, Hughes, N, Ellis, D, Combrink, K, Jarousse, N, Altmann, E. | | Deposit date: | 2014-11-13 | | Release date: | 2015-01-14 | | Last modified: | 2015-01-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-based design and optimization of potent inhibitors of the adenoviral protease.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4WX7
 
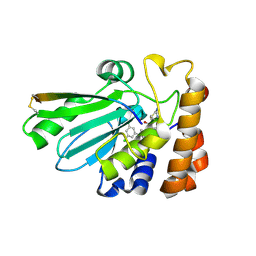 | | Crystal structure of adenovirus 8 protease with a nitrile inhibitor | | Descriptor: | 3-[2-(3,5-dichlorophenyl)-2-methylpropanoyl]-N-(2-{[(2Z)-2-iminoethyl]amino}-2-oxoethyl)-4-methoxybenzamide, PVI, Protease | | Authors: | Grosche, P, Sirockin, F, Mac Sweeney, A, Ramage, P, Erbel, P, Melkko, S, Bernardi, A, Hughes, N, Ellis, D, Combrink, K, Jarousse, N, Altmann, E. | | Deposit date: | 2014-11-13 | | Release date: | 2015-01-14 | | Last modified: | 2015-01-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design and optimization of potent inhibitors of the adenoviral protease.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
6IA2
 
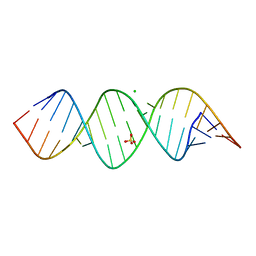 | | Crystal structure of a self-complementary RNA duplex recognized by Com | | Descriptor: | CHLORIDE ION, RNA (5'-R(*AP*GP*AP*GP*AP*AP*CP*CP*CP*GP*GP*AP*GP*UP*UP*CP*CP*CP*U)-3'), SULFATE ION | | Authors: | Nowacka, M, Fernandes, H, Kiliszek, A, Bernat, A, Lach, G, Bujnicki, J.M. | | Deposit date: | 2018-11-26 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Specific interaction of zinc finger protein Com with RNA and the crystal structure of a self-complementary RNA duplex recognized by Com.
Plos One, 14, 2019
|
|
2WBB
 
 | | FRUCTOSE-1,6-BISPHOSPHATASE(D-FRUCTOSE-1,6-BISPHOSPHATE-1- PHOSPHOHYDROLASE) (E.C.3.1.3.11) COMPLEXED WITH AN AMP SITE INHIBITOR | | Descriptor: | FRUCTOSE-1,6-BISPHOSPHATASE 1, N-{[(2Z)-5-BROMO-1,3-THIAZOL-2(3H)-YLIDENE]CARBAMOYL}-4-METHYLBENZENESULFONAMIDE | | Authors: | Ruf, A, Joseph, C, Benz, J, Fol, B, Tetaz, T, Kitas, E, Mohr, P, Kuhn, B, Wessel, H.P, Hebeisen, P, Haap, W, Huber, W, Alvarez Sanchez, R, Paehler, A, Bernadeau, A, Gubler, M, Schott, B, Tozzo, E. | | Deposit date: | 2009-02-26 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Sulfonylureido Thiazoles as Fructose-1,6-Bisphosphatase Inhibitors for the Treatment of Type-2 Diabetes.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2WBD
 
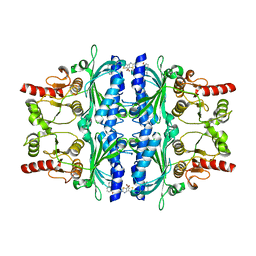 | | FRUCTOSE-1,6-BISPHOSPHATASE(D-FRUCTOSE-1,6-BISPHOSPHATE-1- PHOSPHOHYDROLASE) (E.C.3.1.3.11) COMPLEXED WITH AN AMP SITE INHIBITOR | | Descriptor: | FRUCTOSE-1,6-BISPHOSPHATASE 1, N-[(5-bromo-1,3-thiazol-2-yl)carbamoyl]-3-ethylbenzenesulfonamide | | Authors: | Ruf, A, Joseph, C, Benz, J, Fol, B, Tetaz, T, Kitas, E, Mohr, P, Kuhn, B, Wessel, H.P, Hebeisen, P, Haap, W, Huber, W, Alvarez Sanchez, R, Paehler, A, Bernadeau, A, Gubler, M, Schott, B, Tozzo, E. | | Deposit date: | 2009-02-26 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sulfonylureido Thiazoles as Fructose-1,6-Bisphosphatase Inhibitors for the Treatment of Type-2 Diabetes.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2XR6
 
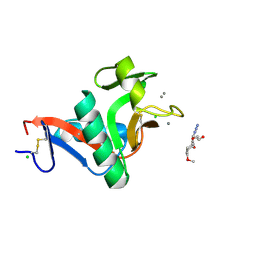 | | Crystal structure of the complex of the carbohydrate recognition domain of human DC-SIGN with pseudo trimannoside mimic. | | Descriptor: | 2-AZIDOETHANOL, CALCIUM ION, CD209 ANTIGEN, ... | | Authors: | Thepaut, M, Suitkeviciute, I, Sattin, S, Reina, J, Bernardi, A, Fieschi, F. | | Deposit date: | 2010-09-10 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Unique Dc-Sign Clustering Activity of a Small Glycomimetic: A Lesson for Ligand Design.
Acs Chem.Biol., 9, 2014
|
|
4PIS
 
 | | Crystal structure of human adenovirus 8 protease in complex with a nitrile inhibitor | | Descriptor: | N~2~-[(2R)-2-(3,5-dichlorophenyl)-2-(dimethylamino)acetyl]-N-({2-[(Z)-iminomethyl]pyrimidin-4-yl}methyl)-L-isoleucinamide, PVI, Protease | | Authors: | Mac Sweeney, A, Grosche, P, Ellis, D, Combrink, K, Erbel, P, Hughes, N, Sirockin, F, Melkko, S, Bernardi, A, Ramage, P, Jarousse, N, Altmann, E. | | Deposit date: | 2014-05-09 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and structure-based optimization of adenain inhibitors.
Acs Med.Chem.Lett., 5, 2014
|
|
4PIE
 
 | | Crystal structure of human adenovirus 2 protease a substrate based nitrile inhibitor | | Descriptor: | ACETATE ION, N-{(2S)-2-(3-chlorophenyl)-2-[(methylsulfonyl)amino]acetyl}-L-phenylalanyl-N-[(2Z)-2-iminoethyl]glycinamide, Pre-protein VI, ... | | Authors: | Mac Sweeney, A, Grosche, P, Ellis, D, Combrink, C, Erbel, C, Hughes, N, Sirockin, F, Melkko, S, Bernardi, A, Ramage, P, Jarousse, N, Altmann, E. | | Deposit date: | 2014-05-08 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery and structure-based optimization of adenain inhibitors.
Acs Med.Chem.Lett., 5, 2014
|
|
4PID
 
 | | Crystal structure of human adenovirus 2 protease with a weak pyrimidine nitrile inhibitor | | Descriptor: | ACETATE ION, N-benzyl-2-[(Z)-iminomethyl]pyrimidine-5-carboxamide, Pre-protein VI, ... | | Authors: | Mac Sweeney, A, Grosche, P, Ellis, D, Combrink, K, Erbel, P, Hughes, N, Sirockin, F, Melkko, S, Bernardi, A, Ramage, P, Jarousse, N, Altmann, E. | | Deposit date: | 2014-05-08 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Discovery and structure-based optimization of adenain inhibitors.
Acs Med.Chem.Lett., 5, 2014
|
|
4PIQ
 
 | | Crystal structure of human adenovirus 8 protease with a nitrile inhibitor | | Descriptor: | N-[(3,5-dichlorophenyl)acetyl]-L-threonyl-N-[(2Z)-2-iminoethyl]glycinamide, PVI, Protease | | Authors: | Mac Sweeney, A, Grosche, P, Ellis, D, Combrink, K, Erbel, P, Hughes, N, Sirockin, F, Melkko, S, Bernardi, A, Ramage, P, Jarousse, N, Altmann, E. | | Deposit date: | 2014-05-09 | | Release date: | 2014-09-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery and structure-based optimization of adenain inhibitors.
Acs Med.Chem.Lett., 5, 2014
|
|
3SZ1
 
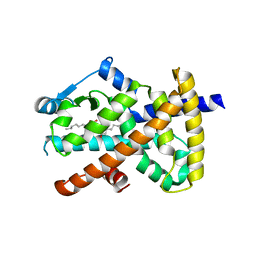 | | Human PPAR gamma ligand binding domain in complex with luteolin and myristic acid | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-chromen-4-one, MYRISTIC ACID, Peroxisome proliferator-activated receptor gamma, ... | | Authors: | Puhl, A.C, Bernardes, A, Polikarpov, I. | | Deposit date: | 2011-07-18 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mode of peroxisome proliferator-activated receptor gamma activation by luteolin.
Mol.Pharmacol., 81, 2012
|
|
5G6U
 
 | | Crystal structure of langerin carbohydrate recognition domain with GlcNS6S | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Porkolab, V, Chabrol, E, Varga, N, Ordanini, S, Sutkeviciute, I, Thepaut, M, Bernardi, A, Fieschi, F. | | Deposit date: | 2016-07-21 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | Rational-Differential Design of Highly Specific Glycomimetic Ligands: Targeting DC-SIGN and Excluding Langerin Recognition.
ACS Chem. Biol., 13, 2018
|
|
6XOF
 
 | | Crystal structure of SCLam, a non-specific endo-beta-1,3(4)-glucanase from family GH16 | | Descriptor: | CALCIUM ION, GH16 family protein, GLYCEROL | | Authors: | Liberato, M.V, Bernardes, A, Polikarpov, I, Squina, F. | | Deposit date: | 2020-07-07 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Insights into the dual cleavage activity of the GH16 laminarinase enzyme class on beta-1,3 and beta-1,4 glycosidic bonds.
J.Biol.Chem., 296, 2021
|
|
