6XCN
 
 | |
6XCM
 
 | |
6OIX
 
 | | Structure of Escherichia coli dGTPase bound to GTP | | Descriptor: | Deoxyguanosinetriphosphate triphosphohydrolase, GUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION | | Authors: | Barnes, C.O, Wu, Y, Calero, G. | | Deposit date: | 2019-04-09 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The crystal structure of dGTPase reveals the molecular basis of dGTP selectivity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5C4X
 
 | | Crystal structure of a transcribing RNA Polymerase II complex reveals a complete transcription bubble | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Barnes, C.O, Calero, M, Malik, I, Spahr, H, Zhang, Q, Pullara, F, Kaplan, C.D, Calero, G. | | Deposit date: | 2015-06-18 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal Structure of a Transcribing RNA Polymerase II Complex Reveals a Complete Transcription Bubble.
Mol.Cell, 59, 2015
|
|
5C4J
 
 | | Crystal structure of a transcribing RNA Polymerase II complex reveals a complete transcription bubble | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Barnes, C.O, Calero, M, Malik, I, Spahr, H, Zhang, Q, Pullara, F, Kaplan, C.D, Calero, G. | | Deposit date: | 2015-06-18 | | Release date: | 2015-08-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal Structure of a Transcribing RNA Polymerase II Complex Reveals a Complete Transcription Bubble.
Mol.Cell, 59, 2015
|
|
5C4A
 
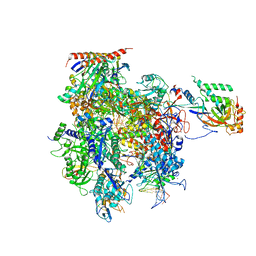 | | Crystal structure of a transcribing RNA Polymerase II complex reveals a complete transcription bubble | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Barnes, C.O, Calero, M, Malik, I, Saphr, H, Zhang, Q, Pullara, F, Kaplan, C.D, Calero, G. | | Deposit date: | 2015-06-17 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Crystal Structure of a Transcribing RNA Polymerase II Complex Reveals a Complete Transcription Bubble.
Mol.Cell, 59, 2015
|
|
5C3E
 
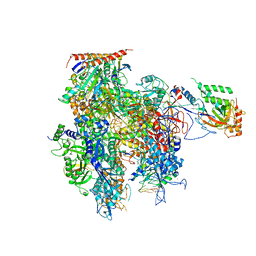 | | Crystal structure of a transcribing RNA Polymerase II complex reveals a complete transcription bubble | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Barnes, C.O, Calero, M, Malik, I, Spahr, H, Zhang, Q, Pullara, F, Kaplan, C.D, Calero, G. | | Deposit date: | 2015-06-17 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal Structure of a Transcribing RNA Polymerase II Complex Reveals a Complete Transcription Bubble.
Mol.Cell, 59, 2015
|
|
5C44
 
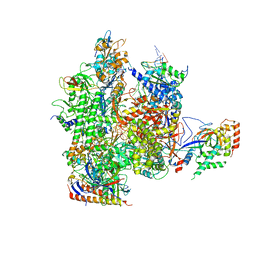 | | Crystal structure of a transcribing RNA Polymerase II complex reveals a complete transcription bubble | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Barnes, C.O, Calero, M, Malik, I, Spahr, H, Zhang, Q, Pullara, F, Kaplan, C.D, Calero, G. | | Deposit date: | 2015-06-17 | | Release date: | 2015-08-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Crystal Structure of a Transcribing RNA Polymerase II Complex Reveals a Complete Transcription Bubble.
Mol.Cell, 59, 2015
|
|
7K8S
 
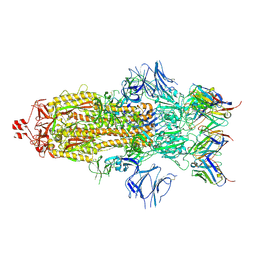 | | Structure of the SARS-CoV-2 S 2P trimer in complex with the human neutralizing antibody Fab fragment, C002 (state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C002 Fab Heavy Chain, ... | | Authors: | Barnes, C.O, Malyutin, A.G, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
6CHB
 
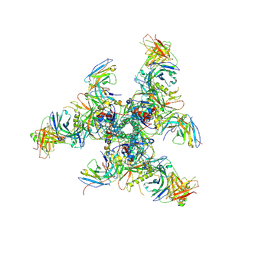 | |
6CH9
 
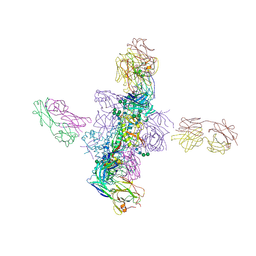 | |
6CH7
 
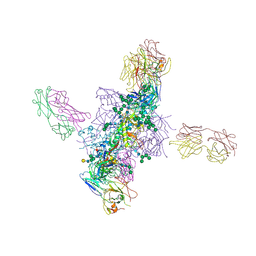 | |
6CH8
 
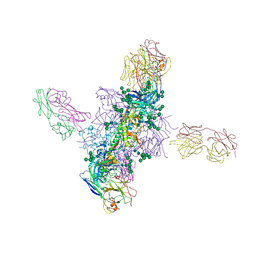 | |
6OIW
 
 | | Structure of Escherichia coli dGTPase bound to dGTP-1-thiol | | Descriptor: | 2'-deoxyguanosine-5'-O-(1-thiotriphosphate), Deoxyguanosinetriphosphate triphosphohydrolase, MAGNESIUM ION, ... | | Authors: | Barnes, C.O, Wu, Y, Calero, G. | | Deposit date: | 2019-04-09 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | The crystal structure of dGTPase reveals the molecular basis of dGTP selectivity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OIY
 
 | | Structure of Escherichia coli bound to dGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Deoxyguanosinetriphosphate triphosphohydrolase, MANGANESE (II) ION | | Authors: | Barnes, C.O, Wu, Y, Calero, G. | | Deposit date: | 2019-04-09 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | The crystal structure of dGTPase reveals the molecular basis of dGTP selectivity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OIV
 
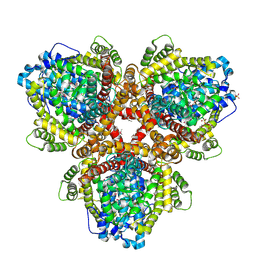 | | XFEL structure of Escherichia coli dGTPase | | Descriptor: | Deoxyguanosinetriphosphate triphosphohydrolase, MANGANESE (II) ION, SULFATE ION | | Authors: | Barnes, C.O, Wu, Y, Calero, G. | | Deposit date: | 2019-04-09 | | Release date: | 2019-06-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | The crystal structure of dGTPase reveals the molecular basis of dGTP selectivity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7UCE
 
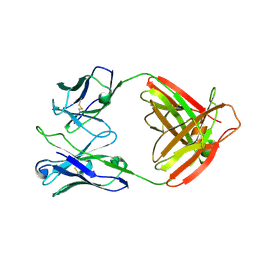 | |
7UCF
 
 | |
7UCG
 
 | |
7K8Z
 
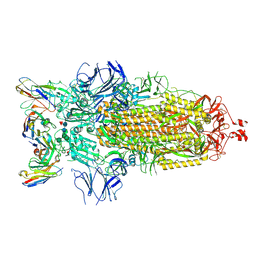 | |
7K8U
 
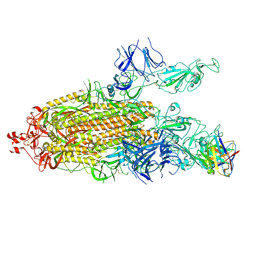 | | Structure of the SARS-CoV-2 S 6P trimer in complex with the human neutralizing antibody Fab fragment, C104 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C104 Fab Heavy Chain, ... | | Authors: | Barnes, C.O, Malyutin, A.G, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K90
 
 | | Structure of the SARS-CoV-2 S 6P trimer in complex with the human neutralizing antibody Fab fragment, C144 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C144 Fab Heavy Chain, C144 Fab Light Chain, ... | | Authors: | Barnes, C.O, Esswein, S.R, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
8E7M
 
 | |
7R8N
 
 | | Structure of the SARS-CoV-2 S 6P trimer in complex with neutralizing antibody C051 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C051 Fab Heavy Chain, ... | | Authors: | Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2021-06-26 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
7R8L
 
 | | Structure of the SARS-CoV-2 RBD in complex with neutralizing antibody C099 and CR3022 | | Descriptor: | C099 Fab Heavy Chain, C099 Fab Light Chain, CR3022 Fab heavy chain, ... | | Authors: | Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2021-06-26 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
