5VHG
 
 | | Crystal structure of pentad mutant GAPR-1 | | Descriptor: | Golgi-associated plant pathogenesis-related protein 1, SULFATE ION | | Authors: | Li, Y, Zhao, Y, Su, M, Chakravarthy, S, Colbert, C.L, Levine, B, Sinha, S.C. | | Deposit date: | 2017-04-13 | | Release date: | 2017-09-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Structural insights into the interaction of the conserved mammalian proteins GAPR-1 and Beclin 1, a key autophagy protein.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5KTQ
 
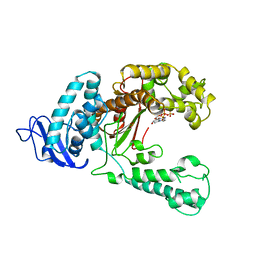 | | LARGE FRAGMENT OF TAQ DNA POLYMERASE BOUND TO DCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, PROTEIN (DNA POLYMERASE I) | | Authors: | Li, Y, Kong, Y, Korolev, S, Waksman, G. | | Deposit date: | 1998-09-22 | | Release date: | 1998-09-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the Klenow fragment of Thermus aquaticus DNA polymerase I complexed with deoxyribonucleoside triphosphates.
Protein Sci., 7, 1998
|
|
5DO2
 
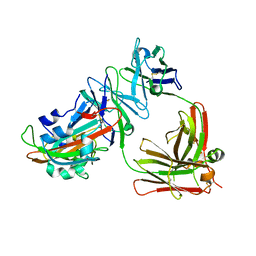 | | Complex structure of MERS-RBD bound with 4C2 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4C2 heavy chain, 4C2 light chain, ... | | Authors: | Li, Y, Wan, Y, Liu, P, Zhao, J, Lu, G, Qi, J, Wang, Q, Lu, X, Wu, Y, Liu, W, Yuen, K.Y, Perlman, S, Gao, G.F, Yan, J. | | Deposit date: | 2015-09-10 | | Release date: | 2015-10-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.409 Å) | | Cite: | A humanized neutralizing antibody against MERS-CoV targeting the receptor-binding domain of the spike protein.
Cell Res., 25, 2015
|
|
3D34
 
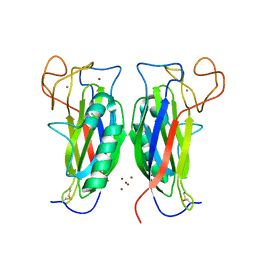 | | Structure of the F-spondin domain of mindin | | Descriptor: | CALCIUM ION, NICKEL (II) ION, Spondin-2 | | Authors: | Li, Y, Mariuzza, R.A. | | Deposit date: | 2008-05-09 | | Release date: | 2009-02-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the F-spondin domain of mindin, an integrin ligand and pattern recognition molecule.
Embo J., 28, 2009
|
|
4G72
 
 | |
4G7Q
 
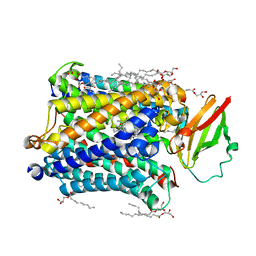 | |
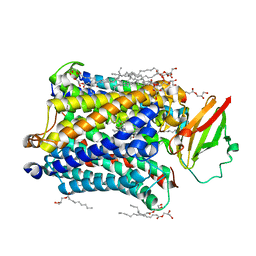
4G71
 
 | |
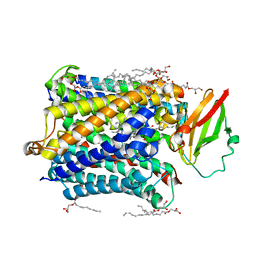
4G7R
 
 | |
7BV6
 
 | | Crystal structure of the autophagic STX17/SNAP29/VAMP8 SNARE complex | | Descriptor: | Synaptosomal-associated protein 29, Syntaxin-17, Vesicle-associated membrane protein 8 | | Authors: | Li, Y, Pan, L.F. | | Deposit date: | 2020-04-09 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Decoding three distinct states of the Syntaxin17 SNARE motif in mediating autophagosome-lysosome fusion.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7BV4
 
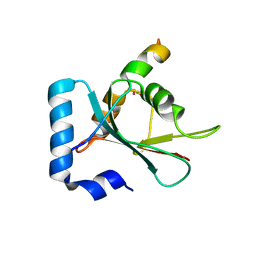 | | Crystal structure of STX17 LIR region in complex with GABARAP | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Gamma-aminobutyric acid receptor-associated protein, ... | | Authors: | Li, Y, Pan, L.F. | | Deposit date: | 2020-04-09 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Decoding three distinct states of the Syntaxin17 SNARE motif in mediating autophagosome-lysosome fusion.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1SL2
 
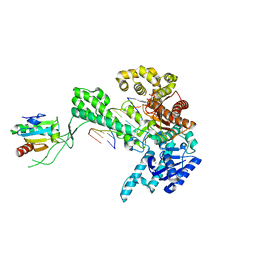 | | Ternary 5' complex of T7 DNA polymerase with a DNA primer/template containing a cis-syn thymine dimer on the template and an incoming nucleotide | | Descriptor: | 2',3'-DIDEOXYADENOSINE-5'-TRIPHOSPHATE, 5'-D(*CP*CP*CP*(TTD)P*AP*GP*GP*CP*AP*CP*TP*GP*GP*CP*CP*GP*TP*CP*GP*TP*TP*TP*TP*CP*G)-3', 5'-D(*CP*GP*AP*AP*AP*AP*CP*GP*AP*CP*GP*GP*CP*CP*AP*GP*TP*GP*CP*CP*TP*(2DA))-3', ... | | Authors: | Li, Y, Dutta, S, Doublie, S, Bdour, H.M, Taylor, J.S, Ellenberger, T. | | Deposit date: | 2004-03-05 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Nucleotide insertion opposite a cis-syn thymine dimer by a replicative DNA polymerase from bacteriophage T7.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1SKW
 
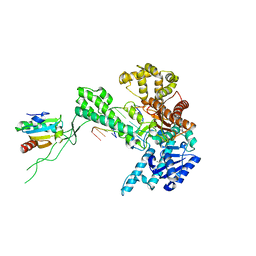 | | Binary 3' complex of T7 DNA polymerase with a DNA primer/template containing a disordered cis-syn thymine dimer on the template | | Descriptor: | 5'-D(*CP*CP*CP*(TTD)P*AP*GP*GP*CP*AP*CP*TP*GP*GP*CP*CP*GP*TP*CP*GP*TP*TP*TP*TP*CP*G)-3', 5'-D(*CP*GP*AP*AP*AP*AP*CP*GP*AP*C*GP*GP*CP*CP*AP*GP*TP*GP*CP*CP*(2DT))-3', DNA polymerase, ... | | Authors: | Li, Y, Dutta, S, Doublie, S, Bdour, H.M, Taylor, J.S, Ellenberger, T. | | Deposit date: | 2004-03-05 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Nucleotide insertion opposite a cis-syn thymine dimer by a replicative DNA polymerase from bacteriophage T7.
Nat.Struct.Mol.Biol., 11, 2004
|
|
4P5A
 
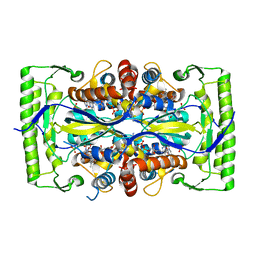 | | Crystal structure of a UMP/dUMP methylase PolB from Streptomyces cacaoi bound with 5-Br UMP | | Descriptor: | 5-BROMO-URIDINE-5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, Thymidylate synthase ThyX | | Authors: | Li, Y, Chen, W, Li, J, Xia, Z, Deng, Z, Zhou, J. | | Deposit date: | 2014-03-15 | | Release date: | 2015-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of a UMP/dUMP methylase PolB from Streptomyces cacaoi with 5-Br UMP
To Be Published
|
|
7SOY
 
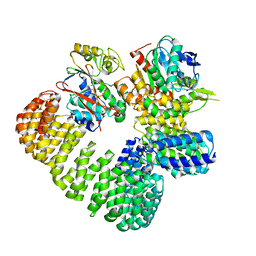 | | The structure of the PP2A-B56gamma1 holoenzyme-PME-1 complex | | Descriptor: | Isoform Gamma-1 of Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform, Protein phosphatase methylesterase 1, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | Authors: | Li, Y, Balakrishnan, V.K, Rowse, M, Novikova, I.V, Xing, Y. | | Deposit date: | 2021-11-01 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Coupling to short linear motifs creates versatile PME-1 activities in PP2A holoenzyme demethylation and inhibition.
Elife, 11, 2022
|
|
8S1R
 
 | |
3CS8
 
 | |
7BRE
 
 | | The crystal structure of MLL2 in complex with ASH2L and RBBP5 | | Descriptor: | Histone-lysine N-methyltransferase 2B, Retinoblastoma-binding protein 5, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Li, Y, Zhao, L, Chen, Y. | | Deposit date: | 2020-03-28 | | Release date: | 2020-07-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Crystal Structure of MLL2 Complex Guides the Identification of a Methylation Site on P53 Catalyzed by KMT2 Family Methyltransferases.
Structure, 28, 2020
|
|
4O8W
 
 | | Crystal Structure of the GerD spore germination protein | | Descriptor: | Spore germination protein | | Authors: | Li, Y, Jin, K, Ghosh, S, Devarakonda, P, Carlson, K, Davis, A, Stewart, K, Cammett, E, Rossi, P.P, Setlow, B, Lu, M, Setlow, P, Hao, B. | | Deposit date: | 2013-12-30 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Structural and Functional Analysis of the GerD Spore Germination Protein of Bacillus Species.
J.Mol.Biol., 426, 2014
|
|
3L6F
 
 | |
5F59
 
 | | The crystal structure of MLL3 SET domain | | Descriptor: | Histone-lysine N-methyltransferase 2C, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Li, Y, Lei, M, Chen, Y. | | Deposit date: | 2015-12-04 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural basis for activity regulation of MLL family methyltransferases.
Nature, 530, 2016
|
|
5F6L
 
 | | The crystal structure of MLL1 (N3861I/Q3867L) in complex with RbBP5 and Ash2L | | Descriptor: | Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Li, Y, Lei, M, Chen, Y. | | Deposit date: | 2015-12-06 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for activity regulation of MLL family methyltransferases.
Nature, 530, 2016
|
|
5F5E
 
 | |
5F6K
 
 | | Crystal structure of the MLL3-Ash2L-RbBP5 complex | | Descriptor: | Histone-lysine N-methyltransferase 2C, Retinoblastoma-binding protein 5, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Li, Y, Lei, M, Chen, Y. | | Deposit date: | 2015-12-06 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.411 Å) | | Cite: | Structural basis for activity regulation of MLL family methyltransferases.
Nature, 530, 2016
|
|
1DTL
 
 | | CRYSTAL STRUCTURE OF CALCIUM-SATURATED (3CA2+) CARDIAC TROPONIN C COMPLEXED WITH THE CALCIUM SENSITIZER BEPRIDIL AT 2.15 A RESOLUTION | | Descriptor: | 1-ISOBUTOXY-2-PYRROLIDINO-3[N-BENZYLANILINO] PROPANE, CALCIUM ION, CARDIAC TROPONIN C | | Authors: | Li, Y, Love, M.L, Putkey, J.A, Cohen, C. | | Deposit date: | 2000-01-12 | | Release date: | 2000-05-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Bepridil opens the regulatory N-terminal lobe of cardiac troponin C.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1HQR
 
 | | CRYSTAL STRUCTURE OF A SUPERANTIGEN BOUND TO THE HIGH-AFFINITY, ZINC-DEPENDENT SITE ON MHC CLASS II | | Descriptor: | HLA-DR ALPHA CHAIN, HLA-DR BETA CHAIN, MYELIN BASIC PROTEIN, ... | | Authors: | Li, Y, Li, H, Dimasi, N, Schlievert, P, Mariuzza, R. | | Deposit date: | 2000-12-19 | | Release date: | 2001-01-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a superantigen bound to the high-affinity, zinc-dependent site on MHC class II.
Immunity, 14, 2001
|
|
