4JFF
 
 | | Preservation of peptide specificity during TCR-MHC contact dominated affinity enhancement of a melanoma-specific TCR | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Madura, F, Sewell, A.K. | | Deposit date: | 2013-02-28 | | Release date: | 2013-05-29 | | Last modified: | 2014-04-02 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | T-cell receptor specificity maintained by altered thermodynamics.
J.Biol.Chem., 288, 2013
|
|
2F53
 
 | | Directed Evolution of Human T-cell Receptor CDR2 residues by phage display dramatically enhances affinity for cognate peptide-MHC without apparent cross-reactivity | | Descriptor: | Beta-2-microglobulin, Cancer/testis antigen 1B, GLYCEROL, ... | | Authors: | Rizkallah, P.J, Jakobsen, B.K, Dunn, S.M, Sami, M. | | Deposit date: | 2005-11-25 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Directed evolution of human T cell receptor CDR2 residues by phage display dramatically enhances affinity for cognate peptide-MHC without increasing apparent cross-reactivity.
Protein Sci., 15, 2006
|
|
2E6Y
 
 | | Covalent complex of orotidine 5'-monophosphate decarboxylase (ODCase) with 6-Iodo-UMP | | Descriptor: | GLYCEROL, Orotidine 5'-phosphate decarboxylase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Fujihashi, M, Bello, A.M, Kotra, L.P, Pai, E.F. | | Deposit date: | 2007-01-05 | | Release date: | 2007-02-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Potent, Covalent Inhibitor of Orotidine 5'-Monophosphate Decarboxylase with Antimalarial Activity.
J.Med.Chem., 50, 2007
|
|
2FNI
 
 | | PseC aminotransferase involved in pseudoaminic acid biosynthesis | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, aminotransferase | | Authors: | Cygler, M, Matte, A, Lunin, V.V, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-01-11 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Functional Characterization of PseC, an Aminotransferase Involved in the Biosynthesis of Pseudaminic Acid, an Essential Flagellar Modification in Helicobacter pylori
J.Biol.Chem., 281, 2006
|
|
2FNU
 
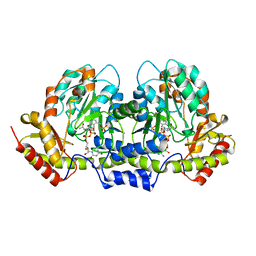 | | PseC aminotransferase with external aldimine | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE, aminotransferase | | Authors: | Cygler, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-01-11 | | Release date: | 2006-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Characterization of PseC, an Aminotransferase Involved in the Biosynthesis of Pseudaminic Acid, an Essential Flagellar Modification in Helicobacter pylori
J.Biol.Chem., 281, 2006
|
|
2FN6
 
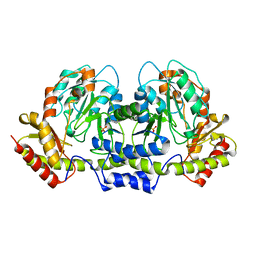 | | Helicobacter pylori PseC, aminotransferase involved in the biosynthesis of pseudoaminic acid | | Descriptor: | AMINOTRANSFERASE, PHOSPHATE ION | | Authors: | Cygler, M, Lunin, V.V, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-01-10 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.483 Å) | | Cite: | Structural and Functional Characterization of PseC, an Aminotransferase Involved in the Biosynthesis of Pseudaminic Acid, an Essential Flagellar Modification in Helicobacter pylori
J.Biol.Chem., 281, 2006
|
|
4JFQ
 
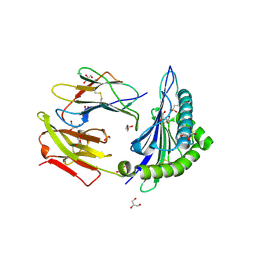 | | A2 HLA complex with L8A heteroclitic variant of Melanoma peptide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Madura, F, Sewell, A.K, Baker, B.M. | | Deposit date: | 2013-02-28 | | Release date: | 2013-05-29 | | Last modified: | 2013-07-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | T-cell receptor specificity maintained by altered thermodynamics.
J.Biol.Chem., 288, 2013
|
|
5YEV
 
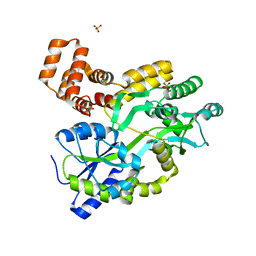 | | Murine DR3 death domain | | Descriptor: | SULFATE ION, TNFRSF25 death domain | | Authors: | Yin, X, Jin, T. | | Deposit date: | 2017-09-19 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and activation mechanism of DR3 death domain.
Febs J., 286, 2019
|
|
5YGP
 
 | | Human TNFRSF25 death domain mutant-D412E | | Descriptor: | SULFATE ION, TNFRSF25 death domain, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yin, X, Jin, T.C. | | Deposit date: | 2017-09-25 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure and activation mechanism of DR3 death domain.
Febs J., 286, 2019
|
|
5YBJ
 
 | | Structure of apo KANK1 ankyrin domain | | Descriptor: | GLYCEROL, KN motif and ankyrin repeat domain-containing protein 1 | | Authors: | Guo, Q, Liao, S, Min, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-05 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | Structural basis for the recognition of kinesin family member 21A (KIF21A) by the ankyrin domains of KANK1 and KANK2 proteins.
J. Biol. Chem., 293, 2018
|
|
5YBV
 
 | | The structure of the KANK2 ankyrin domain with the KIF21A peptide | | Descriptor: | GLYCEROL, KN motif and ankyrin repeat domain-containing protein 2, Kinesin-like protein KIF21A, ... | | Authors: | Guo, Q, Liao, S, Min, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-05 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural basis for the recognition of kinesin family member 21A (KIF21A) by the ankyrin domains of KANK1 and KANK2 proteins.
J. Biol. Chem., 293, 2018
|
|
8IF5
 
 | | AFB1-AF26 APTAMER COMPLEX | | Descriptor: | AFB1 DNA aptamer (26-MER), AFLATOXIN B1 | | Authors: | Xu, G.H, Wang, C, Li, C.G. | | Deposit date: | 2023-02-17 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for high-affinity recognition of aflatoxin B1 by a DNA aptamer.
Nucleic Acids Res., 51, 2023
|
|
4MNQ
 
 | | TCR-peptide specificity overrides affinity enhancing TCR-MHC interactions | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Sewell, A.K, Jakobsen, B.K. | | Deposit date: | 2013-09-11 | | Release date: | 2013-11-13 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (2.742 Å) | | Cite: | T-cell receptor (TCR)-peptide specificity overrides affinity-enhancing TCR-major histocompatibility complex interactions.
J.Biol.Chem., 289, 2014
|
|
5YBU
 
 | | Structure of the KANK1 ankyrin domain in complex with KIF21A peptide | | Descriptor: | KN motif and ankyrin repeat domain-containing protein 1, Kinesin-like protein KIF21A | | Authors: | Guo, Q, Liao, S, Min, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-05 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for the recognition of kinesin family member 21A (KIF21A) by the ankyrin domains of KANK1 and KANK2 proteins.
J. Biol. Chem., 293, 2018
|
|
4M7R
 
 | |
4JFE
 
 | | Preservation of peptide specificity during TCR-MHC contact dominated affinity enhancement of a melanoma-specific TCR | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Madura, F, Sewell, A.K. | | Deposit date: | 2013-02-28 | | Release date: | 2013-05-29 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | T-cell receptor specificity maintained by altered thermodynamics.
J.Biol.Chem., 288, 2013
|
|
5YGS
 
 | | Human TNFRSF25 death domain | | Descriptor: | Human TNRSF25 death domain, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yin, X, Jin, T. | | Deposit date: | 2017-09-26 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.691 Å) | | Cite: | Crystal structure and activation mechanism of DR3 death domain.
Febs J., 286, 2019
|
|
5W0E
 
 | | CREBBP bromodomain in complex with Cpd19 (3-(7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl)-N-methyl-1-(tetrahydro-2H-pyran-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carboxamide) | | Descriptor: | 3-[7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-N-methyl-1-(oxan-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carboxamide, CREB-binding protein | | Authors: | Murray, J.M. | | Deposit date: | 2017-05-30 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | A Unique Approach to Design Potent and Selective Cyclic Adenosine Monophosphate Response Element Binding Protein, Binding Protein (CBP) Inhibitors.
J. Med. Chem., 60, 2017
|
|
4KTD
 
 | | Fab fragment of HIV vaccine-elicited CD4bs-directed antibody, GE136, from non-human primate | | Descriptor: | GE136 Heavy Chain Fab, GE136 Light Chain Fab, GLYCEROL, ... | | Authors: | Poulsen, C, Tran, K, Standfield, R, Wyatt, R.T. | | Deposit date: | 2013-05-20 | | Release date: | 2014-02-05 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Vaccine-elicited primate antibodies use a distinct approach to the HIV-1 primary receptor binding site informing vaccine redesign.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7FAJ
 
 | | CARM1 bound with compound 43 | | Descriptor: | Histone-arginine methyltransferase CARM1, N'-[[3-[4-(3,5-dimethyl-1,2-oxazol-4-yl)-5-methyl-6-phenylazanyl-pyrimidin-2-yl]phenyl]methyl]-N-methyl-ethane-1,2-diamine | | Authors: | Cao, D.Y, Li, J, Xiong, B. | | Deposit date: | 2021-07-06 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2450726 Å) | | Cite: | Structure-Based Discovery of Potent CARM1 Inhibitors for Solid Tumor and Cancer Immunology Therapy.
J.Med.Chem., 64, 2021
|
|
7FAI
 
 | | CARM1 bound with compound 9 | | Descriptor: | Histone-arginine methyltransferase CARM1, N'-[[3-[4-(3,5-dimethyl-1,2-oxazol-4-yl)-5-methyl-6-(oxan-4-ylamino)pyrimidin-2-yl]phenyl]methyl]-N-methyl-ethane-1,2-diamine | | Authors: | Cao, D.Y, Li, J, Xiong, B. | | Deposit date: | 2021-07-06 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09749269 Å) | | Cite: | Structure-Based Discovery of Potent CARM1 Inhibitors for Solid Tumor and Cancer Immunology Therapy.
J.Med.Chem., 64, 2021
|
|
4KTE
 
 | | Fab fragment of HIV vaccine-elicited CD4bs-directed antibody, GE148, from non-human primate | | Descriptor: | GE148 Heavy Chain Fab, GE148 Light Chain Fab, GLYCEROL, ... | | Authors: | Poulsen, C, Tran, K, Stanfield, R, Wyatt, R.T. | | Deposit date: | 2013-05-20 | | Release date: | 2014-02-05 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Vaccine-elicited primate antibodies use a distinct approach to the HIV-1 primary receptor binding site informing vaccine redesign.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5YVF
 
 | | Crystal structure of BFA1 | | Descriptor: | BFA1 | | Authors: | Pu, H, Zhang, L, Duan, Z.K, Peng, L.W, Liu, L. | | Deposit date: | 2017-11-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Nucleus-Encoded Protein BFA1 Promotes Efficient Assembly of the Chloroplast ATP Synthase Coupling Factor 1.
Plant Cell, 30, 2018
|
|
4NW3
 
 | | Crystal structure of MLL CXXC domain in complex with a CpG DNA | | Descriptor: | 5'-D(*GP*CP*CP*AP*TP*CP*GP*AP*TP*GP*GP*C)-3', Histone-lysine N-methyltransferase 2A, ZINC ION | | Authors: | Bian, C, Tempel, W, Chao, X, Walker, J.R, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-05 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | DNA Sequence Recognition of Human CXXC Domains and Their Structural Determinants.
Structure, 26, 2018
|
|
4O64
 
 | | Zinc fingers of KDM2B | | Descriptor: | Lysine-specific demethylase 2B, UNKNOWN ATOM OR ION, ZINC ION | | Authors: | Liu, K, Xu, C, Tempel, W, Walker, J.R, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-20 | | Release date: | 2014-04-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | DNA Sequence Recognition of Human CXXC Domains and Their Structural Determinants.
Structure, 26, 2018
|
|
