8DK3
 
 | | CryoEM structure of Pseudomonas aeruginosa PA14 JetC ATPase domain bound to DNA and cWHD domain of JetA | | Descriptor: | DNA (26-MER), JetA, JetC, ... | | Authors: | Deep, A, Gu, Y, Gao, Y, Ego, K, Herzik, M, Zhou, H, Corbett, K. | | Deposit date: | 2022-07-01 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | The SMC-family Wadjet complex protects bacteria from plasmid transformation by recognition and cleavage of closed-circular DNA.
Mol.Cell, 82, 2022
|
|
8DK1
 
 | | CryoEM structure of JetABC (head construct) from Pseudomonas aeruginosa PA14 | | Descriptor: | JetA, JetB, JetC | | Authors: | Deep, A, Gu, Y, Gao, Y, Ego, K, Herzik, M, Zhou, H, Corbett, K. | | Deposit date: | 2022-07-01 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | The SMC-family Wadjet complex protects bacteria from plasmid transformation by recognition and cleavage of closed-circular DNA.
Mol.Cell, 82, 2022
|
|
1Y5W
 
 | | tRNA-guanine Transglycosylase (TGT) in complex with 6-Amino-4-[2-(4-methylphenyl)ethyl]-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-AMINO-4-[2-(4-METHYLPHENYL)ETHYL]-1,7-DIHYDRO-8H-IMIDAZO[4,5-G]QUINAZOLIN-8-ONE, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Stengl, B, Meyer, E.A, Heine, A, Brenk, R, Diederich, F, Klebe, G. | | Deposit date: | 2004-12-03 | | Release date: | 2005-12-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structures of tRNA-guanine transglycosylase (TGT) in complex with novel and potent inhibitors unravel pronounced induced-fit adaptations and suggest dimer formation upon substrate binding
J.Mol.Biol., 370, 2007
|
|
8DH7
 
 | |
8DDH
 
 | |
8DDF
 
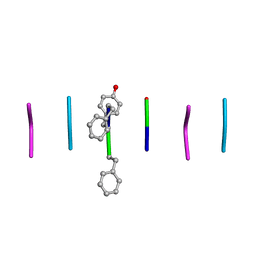 | | Quasi-racemic mixture of L-FWF and D-FYF peptide reveals rippled beta-sheet | | Descriptor: | 1,1,1,3,3,3-hexafluoropropan-2-ol, DPN-DTY-DPN, PHE-TRP-PHE | | Authors: | Sawaya, M.R, Hazari, A, Eisenberg, D.E. | | Deposit date: | 2022-06-18 | | Release date: | 2022-09-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The rippled beta-sheet layer configuration-a novel supramolecular architecture based on predictions by Pauling and Corey.
Chem Sci, 13, 2022
|
|
1JS9
 
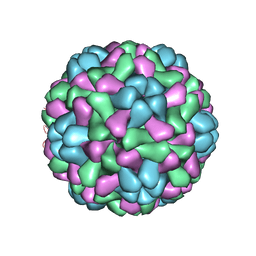 | | Brome Mosaic Virus | | Descriptor: | Coat protein, HEXAETHYLENE GLYCOL, MAGNESIUM ION | | Authors: | Lucas, R.W, Larson, S.B, McPherson, A. | | Deposit date: | 2001-08-16 | | Release date: | 2002-04-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The crystallographic structure of brome mosaic virus.
J.Mol.Biol., 317, 2002
|
|
8EOI
 
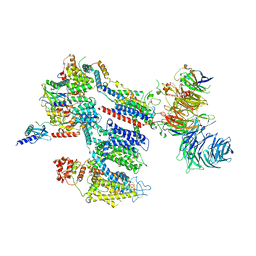 | | Structure of a human EMC:human Cav1.2 channel complex in GDN detergent | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, Z, Mondal, A, Abderemane-Ali, F, Minor, D.L. | | Deposit date: | 2022-10-03 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | EMC chaperone-Ca V structure reveals an ion channel assembly intermediate.
Nature, 619, 2023
|
|
1GSJ
 
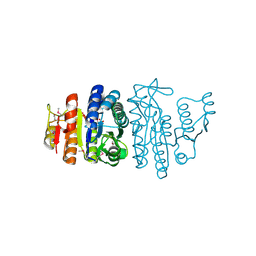 | | Selenomethionine substituted N-acetyl-L-glutamate kinase from Escherichia coli complexed with its substrate n-acetyl-L-glutamate and its substrate analog AMPPNP | | Descriptor: | ACETYLGLUTAMATE KINASE, MAGNESIUM ION, N-ACETYL-L-GLUTAMATE, ... | | Authors: | Ramon-Maiques, S, Marina, A, Gil-Ortiz, F, Fita, I, Rubio, V. | | Deposit date: | 2002-01-07 | | Release date: | 2002-05-16 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of Acetylglutamate Kinase, a Key Enzyme for Arginine Biosynthesis and a Prototype for the Amino Acid Kinase Enzyme Family, During Catalysis
Structure, 10, 2002
|
|
1HD6
 
 | | PHEROMONE ER-22, NMR | | Descriptor: | PHEROMONE ER-22 | | Authors: | Luginbuhl, P, Liu, A, Zerbe, O, Ortenzi, C, Luporini, P, Wuthrich, K. | | Deposit date: | 2000-11-09 | | Release date: | 2000-12-10 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Pheromone Er-22 from Euplotes Raikovi
J.Biomol.NMR, 19, 2001
|
|
8EIT
 
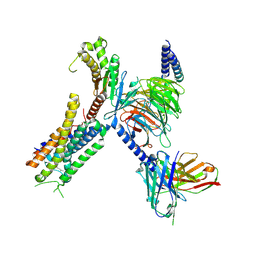 | | Structure of FFAR1-Gq complex bound to DHA | | Descriptor: | A modified Guanine nucleotide-binding protein G(q) subunit alpha, DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Free fatty acid receptor 1, ... | | Authors: | Kumari, P, Inoue, A, Chapman, K, Lian, P, Rosenbaum, D.M. | | Deposit date: | 2022-09-15 | | Release date: | 2023-05-24 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular mechanism of fatty acid activation of FFAR1.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EJK
 
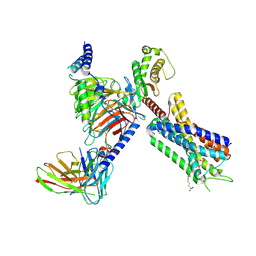 | | Structure of FFAR1-Gq complex bound to TAK-875 in a lipid nanodisc | | Descriptor: | A modified Guanine nucleotide-binding protein G(q) subunit alpha, Free fatty acid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kumari, P, Inoue, A, Chapman, K, Lian, P, Rosenbaum, D.M. | | Deposit date: | 2022-09-17 | | Release date: | 2023-05-24 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular mechanism of fatty acid activation of FFAR1.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EJC
 
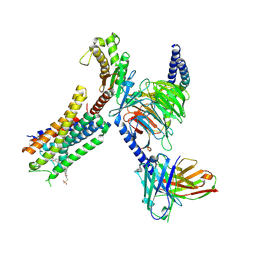 | | Structure of FFAR1-Gq complex bound to TAK-875 | | Descriptor: | A modified Guanine nucleotide-binding protein G(q) subunit alpha, Free fatty acid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kumari, P, Inoue, A, Chapman, K, Lian, P, Rosenbaum, D.M. | | Deposit date: | 2022-09-16 | | Release date: | 2023-05-24 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular mechanism of fatty acid activation of FFAR1.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1GZ9
 
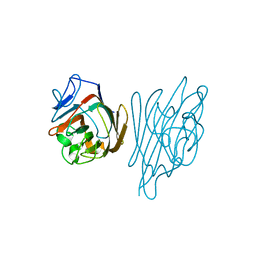 | | High-Resolution Crystal Structure of Erythrina cristagalli Lectin in Complex with 2'-alpha-L-Fucosyllactose | | Descriptor: | CALCIUM ION, ERYTHRINA CRISTA-GALLI LECTIN, MANGANESE (II) ION, ... | | Authors: | Svensson, C, Teneberg, S, Nilsson, C.L, Kjellberg, A, Schwarz, F.P, Sharon, N, Krengel, U. | | Deposit date: | 2002-05-17 | | Release date: | 2002-06-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-Resolution Crystal Structures of Erythrina Cristagalli Lectin in Complex with Lactose and 2'-Alpha-L-Fucosyllactose and Correlation with Thermodynamic Binding Data
J.Mol.Biol., 321, 2002
|
|
8EOG
 
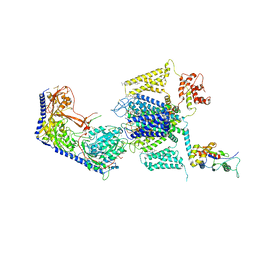 | | Structure of the human L-type voltage-gated calcium channel Cav1.2 complexed with L-leucine | | Descriptor: | (2R)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(dodecanoyloxy)propyl dodecanoate, (2R)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(dodecanoyloxy)propyl heptadecanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, Z, Mondal, A, Abderemane-Ali, F, Minor, D.L. | | Deposit date: | 2022-10-03 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | EMC chaperone-Ca V structure reveals an ion channel assembly intermediate.
Nature, 619, 2023
|
|
1H1B
 
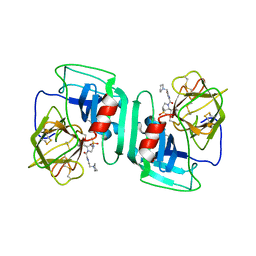 | | Crystal structure of human neutrophil elastase complexed with an inhibitor (GW475151) | | Descriptor: | (2S)-3-METHYL-2-((2R,3S)-3-[(METHYLSULFONYL)AMINO]-1-{[2-(PYRROLIDIN-1-YLMETHYL)-1,3-OXAZOL-4-YL]CARBONYL}PYRROLIDIN-2-YL)BUTANOIC ACID, LEUKOCYTE ELASTASE, alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Macdonald, S.J.F, Dowle, M.D, Harrison, L.A, Clarke, G.D.E, Inglis, G.G.A, Johnson, M.R, Smith, R.A, Amour, A, Fleetwood, G, Humphreys, D.C, Molloy, C.R, Dixon, M, Godward, R.E, Wonacott, A.J, Singh, O.M.P, Hodgson, S.T, Hardy, G.W. | | Deposit date: | 2002-07-05 | | Release date: | 2002-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Further Pyrrolidine Trans-Lactams as Inhibitors of Human Neutrophil Elastase (Hne) with Potential as Development Candidates and the Crystal Structure of Hne Complexed with an Inhibitor (Gw475151)
J.Med.Chem., 45, 2002
|
|
1GT0
 
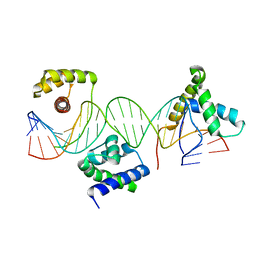 | | Crystal structure of a POU/HMG/DNA ternary complex | | Descriptor: | 5'-D(*AP*TP*CP*CP*CP*AP*TP*TP*AP*GP* CP*AP*TP*CP*CP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3', 5'-D(*TP*TP*CP*TP*TP*TP*GP*TP*TP*TP* GP*GP*AP* TP*GP*CP*TP*AP*AP*TP*GP*GP*GP*A)-3', OCTAMER-BINDING TRANSCRIPTION FACTOR 1, ... | | Authors: | Remenyi, A, Wilmanns, M. | | Deposit date: | 2002-01-09 | | Release date: | 2003-01-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of a POU/Hmg/DNA Ternary Complex Suggests Differential Assembly of Oct4 and Sox2 on Two Enhancers
Genes Dev., 17, 2003
|
|
2Q1L
 
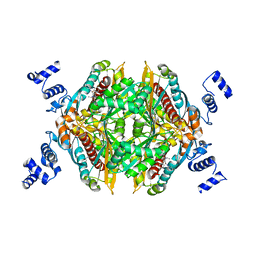 | | Design and Synthesis of Pyrrole-based, Hepatoselective HMG-CoA Reductase Inhibitors | | Descriptor: | (3R,5R)-7-[5-(ANILINOCARBONYL)-3,4-BIS(4-FLUOROPHENYL)-1-ISOPROPYL-1H-PYRROL-2-YL]-3,5-DIHYDROXYHEPTANOIC ACID, 3-hydroxy-3-methylglutaryl-coenzyme A reductase | | Authors: | Pavlovsky, A, Pfefferkorn, J.A, Harris, M.S, Finzel, B.C. | | Deposit date: | 2007-05-24 | | Release date: | 2007-07-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Design and synthesis of hepatoselective, pyrrole-based HMG-CoA reductase inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
1H75
 
 | | Structural basis for the thioredoxin-like activity profile of the glutaredoxin-like protein NrdH-redoxin from Escherichia coli. | | Descriptor: | GLUTAREDOXIN-LIKE PROTEIN NRDH | | Authors: | Stehr, M, Schneider, G, Aslund, F, Holmgren, A, Lindqvist, Y. | | Deposit date: | 2001-07-03 | | Release date: | 2001-08-09 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the Thioredoxin-Like Activity Profile of the Glutaredoxin-Like Nrdh-Redoxin from Escherichia Coli
J.Biol.Chem., 276, 2001
|
|
1H7C
 
 | | human tubulin chaperone cofactor a | | Descriptor: | ACETIC ACID, SULFATE ION, TUBULIN-SPECIFIC CHAPERONE A | | Authors: | Guasch, A, Aloria, K, Perez, R, Campo, R, Avila, J, Zabala, J.C, Coll, M. | | Deposit date: | 2001-07-04 | | Release date: | 2002-06-13 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-Dimensional Structure of Human Tubulin Chaperone Cofactor A
J.Mol.Biol., 318, 2002
|
|
8F5D
 
 | | Architecture of the MurE-MurF ligase bacterial cell wall biosynthesis complex | | Descriptor: | Multifunctional fusion protein, SULFATE ION | | Authors: | Shirakawa, K.T, Sala, F.A, Miyachiro, M.M, Job, V, Trindade, D.M, Dessen, A. | | Deposit date: | 2022-11-14 | | Release date: | 2023-06-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Architecture and genomic arrangement of the MurE-MurF bacterial cell wall biosynthesis complex.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1Y5V
 
 | | tRNA-Guanine Transglycosylase (TGT) in complex with 6-Amino-4-(2-phenylethyl)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-AMINO-4-(2-PHENYLETHYL)-1,7-DIHYDRO-8H-IMIDAZO[4,5-G]QUINAZOLIN-8-ONE, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Stengl, B, Meyer, E.A, Heine, A, Brenk, R, Diederich, F, Klebe, G. | | Deposit date: | 2004-12-03 | | Release date: | 2005-12-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structures of tRNA-guanine transglycosylase (TGT) in complex with novel and potent inhibitors unravel pronounced induced-fit adaptations and suggest dimer formation upon substrate binding
J.Mol.Biol., 370, 2007
|
|
1QA2
 
 | | TAILSPIKE PROTEIN, MUTANT A334V | | Descriptor: | TAILSPIKE PROTEIN | | Authors: | Baxa, U, Steinbacher, S, Weintraub, A, Huber, R, Seckler, R. | | Deposit date: | 1999-04-10 | | Release date: | 2000-01-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutations improving the folding of phage P22 tailspike protein affect its receptor binding activity.
J.Mol.Biol., 293, 1999
|
|
1Y8D
 
 | | Dimeric parallel-stranded tetraplex with 3+1 5' G-tetrad interface, single-residue chain reversal loops and GAG triad in the context of A(GGGG) pentad | | Descriptor: | 5'-D(*GP*GP*GP*GP*TP*GP*GP*GP*AP*GP*GP*AP*GP*GP*GP*T)-3' | | Authors: | Phan, A.T, Kuryavyi, V.V, Ma, J.-B, Faure, A, Andreola, M.-L, Patel, D.J. | | Deposit date: | 2004-12-11 | | Release date: | 2005-02-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An interlocked dimeric parallel-stranded DNA quadruplex: A potent inhibitor of HIV-1 integrase
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1H97
 
 | | Trematode hemoglobin from Paramphistomum epiclitum | | Descriptor: | Globin-3, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Pesce, A, Dewilde, S, Kiger, L, Milani, M, Ascenzi, P, Marden, M.C, Van Hauwaert, M.L, Vanfleteren, J, Moens, L, Bolognesi, M. | | Deposit date: | 2001-03-02 | | Release date: | 2001-06-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Very High Resolution Structure of a Trematode Hemoglobin Displaying a Tyrb10-Tyre7 Heme Distal Residue Pair and High Oxygen Affinity
J.Mol.Biol., 309, 2001
|
|
