8I1I
 
 | | Crystal structure of human MTH1(G2K/D120N mutant) in complex with 2-oxo-dATP at pH 7.7 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Protonation states of Asp residues in the human Nudix hydrolase MTH1 contribute to its broad substrate recognition.
Febs Lett., 597, 2023
|
|
8I1D
 
 | | Crystal structure of human MTH1(G2K mutant) in complex with 2-oxo-dATP at pH 7.7 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Protonation states of Asp residues in the human Nudix hydrolase MTH1 contribute to its broad substrate recognition.
Febs Lett., 597, 2023
|
|
8I1E
 
 | | Crystal structure of human MTH1(G2K mutant) in complex with 2-oxo-dATP at pH 8.0 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Protonation states of Asp residues in the human Nudix hydrolase MTH1 contribute to its broad substrate recognition.
Febs Lett., 597, 2023
|
|
8I18
 
 | |
8I1H
 
 | |
8I1J
 
 | | Crystal structure of human MTH1(G2K/D120N mutant) in complex with 2-oxo-dATP at pH 9.7 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Protonation states of Asp residues in the human Nudix hydrolase MTH1 contribute to its broad substrate recognition.
Febs Lett., 597, 2023
|
|
8I1G
 
 | |
8I19
 
 | |
8I8T
 
 | |
8I1F
 
 | | Crystal structure of human MTH1(G2K mutant) in complex with 2-oxo-dATP at pH 8.6 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Protonation states of Asp residues in the human Nudix hydrolase MTH1 contribute to its broad substrate recognition.
Febs Lett., 597, 2023
|
|
8I8S
 
 | |
2DOO
 
 | | The structure of IMP-1 complexed with the detecting reagent (DansylC4SH) by a fluorescent probe | | Descriptor: | BETA-LACTAMASE IMP-1, N-[4-({[5-(DIMETHYLAMINO)-1-NAPHTHYL]SULFONYL}AMINO)BUTYL]-3-SULFANYLPROPANAMIDE, ZINC ION | | Authors: | Kurosaki, H, Yamaguchi, Y, Yasuzawa, H, Jin, W, Yamagata, Y, Arakawa, Y. | | Deposit date: | 2006-05-01 | | Release date: | 2006-11-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Probing, inhibition, and crystallographic characterization of metallo-beta-lactamase (IMP-1) with fluorescent agents containing dansyl and thiol groups
Chemmedchem, 1, 2006
|
|
1WSF
 
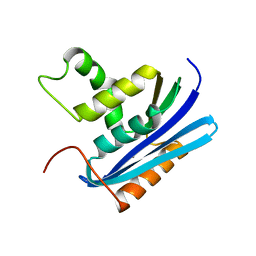 | | Co-crystal structure of E.coli RNase HI active site mutant (D134A*) with Mn2+ | | Descriptor: | MANGANESE (II) ION, Ribonuclease HI | | Authors: | Tsunaka, Y, Takano, K, Matsumura, H, Yamagata, Y, Kanaya, S. | | Deposit date: | 2004-11-05 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of Single Mn(2+) Binding Sites Required for Activation of the Mutant Proteins of E.coli RNase HI at Glu48 and/or Asp134 by X-ray Crystallography
J.Mol.Biol., 345, 2005
|
|
1WSE
 
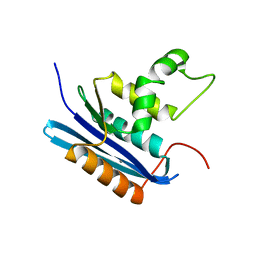 | | Co-crystal structure of E.coli RNase HI active site mutant (E48A*) with Mn2+ | | Descriptor: | MANGANESE (II) ION, Ribonuclease HI | | Authors: | Tsunaka, Y, Takano, K, Matsumura, H, Yamagata, Y, Kanaya, S. | | Deposit date: | 2004-11-05 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of Single Mn(2+) Binding Sites Required for Activation of the Mutant Proteins of E.coli RNase HI at Glu48 and/or Asp134 by X-ray Crystallography
J.Mol.Biol., 345, 2005
|
|
1WSG
 
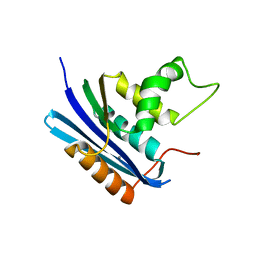 | | Co-crystal structure of E.coli RNase HI active site mutant (E48A/D134N*) with Mn2+ | | Descriptor: | MANGANESE (II) ION, Ribonuclease HI | | Authors: | Tsunaka, Y, Takano, K, Matsumura, H, Yamagata, Y, Kanaya, S. | | Deposit date: | 2004-11-05 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of Single Mn(2+) Binding Sites Required for Activation of the Mutant Proteins of E.coli RNase HI at Glu48 and/or Asp134 by X-ray Crystallography
J.Mol.Biol., 345, 2005
|
|
2ZJC
 
 | | TNFR1 selectve TNF mutant; R1-6 | | Descriptor: | GLYCEROL, Tumor necrosis factor | | Authors: | Mukai, Y, Yamagata, Y, Tsutsumi, Y. | | Deposit date: | 2008-03-05 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Function Relationship of Tumor Necrosis Factor (TNF) and Its Receptor Interaction Based on 3D Structural Analysis of a Fully Active TNFR1-Selective TNF Mutant
J.Mol.Biol., 385, 2009
|
|
2ZPX
 
 | |
3A4F
 
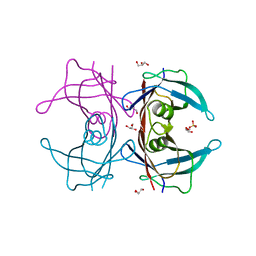 | | Crystal Structure of Human Transthyretin (E54K) | | Descriptor: | GLYCEROL, SULFATE ION, Transthyretin | | Authors: | Miyata, M, Sato, T, Nakamura, T, Ikemizu, S, Yamagata, Y, Kai, H. | | Deposit date: | 2009-07-06 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Role of the glutamic acid 54 residue in transthyretin stability and thyroxine binding
Biochemistry, 49, 2010
|
|
2ZJ9
 
 | | X-ray crystal structure of AmpC beta-Lactamase (AmpC(D)) from an Escherichia coli with a Tripeptide Deletion (Gly286 Ser287 Asp288) on the H10 Helix | | Descriptor: | AmpC, ISOPROPYL ALCOHOL, SODIUM ION | | Authors: | Yamaguchi, Y, Sato, G, Yamagata, Y, Wachino, J, Arakawa, Y, Kurosaki, H. | | Deposit date: | 2008-02-29 | | Release date: | 2009-03-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of AmpC beta-lactamase (AmpCD) from an Escherichia coli clinical isolate with a tripeptide deletion (Gly286-Ser287-Asp288) in the H10 helix
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3ALQ
 
 | | Crystal structure of TNF-TNFR2 complex | | Descriptor: | COBALT (II) ION, Tumor necrosis factor, Tumor necrosis factor receptor superfamily member 1B | | Authors: | Mukai, Y, Nakamura, T, Yamagata, Y, Tsutsumi, Y. | | Deposit date: | 2010-08-06 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Solution of the Structure of the TNF-TNFR2 Complex
Sci.Signal., 3, 2010
|
|
3A4E
 
 | | Crystal structure of Human Transthyretin (E54G) | | Descriptor: | GLYCEROL, SULFATE ION, Transthyretin | | Authors: | Miyata, M, Sato, T, Nakamura, T, Ikemizu, S, Yamagata, Y, Kai, H. | | Deposit date: | 2009-07-06 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Role of the glutamic acid 54 residue in transthyretin stability and thyroxine binding
Biochemistry, 49, 2010
|
|
1X10
 
 | | Structure of Mutant Pyrrolidone Carboxyl Peptidase (E192A) from a Hyperthermophile, Pyrococcus furiosus | | Descriptor: | Pyrrolidone-carboxylate peptidase | | Authors: | Kaushik, J.K, Yamagata, Y, Ogasahara, K, Yutani, K. | | Deposit date: | 2005-03-31 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Completely buried, non-ion-paired glutamic acid contributes favorably to the conformational stability of pyrrolidone carboxyl peptidases from hyperthermophiles.
Biochemistry, 45, 2006
|
|
1Z8X
 
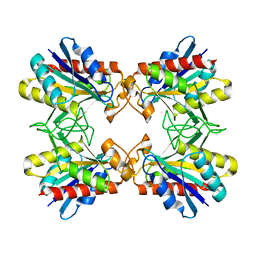 | | Structure of Mutant Pyrrolidone Carboxyl Peptidase (E192V) from a Hyperthermophile, Pyrococcus furiosus | | Descriptor: | Pyrrolidone-carboxylate peptidase | | Authors: | Kaushik, J.K, Yamagata, Y, Ogasahara, K, Yutani, K. | | Deposit date: | 2005-03-31 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Completely buried, non-ion-paired glutamic acid contributes favorably to the conformational stability of pyrrolidone carboxyl peptidases from hyperthermophiles.
Biochemistry, 45, 2006
|
|
1Z8W
 
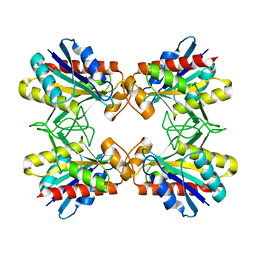 | | Structure of Mutant Pyrrolidone Carboxyl Peptidase (E192I) from a Hyperthermophile, Pyrococcus furiosus | | Descriptor: | Pyrrolidone-carboxylate peptidase | | Authors: | Kaushik, J.K, Yamagata, Y, Ogasahara, K, Yutani, K. | | Deposit date: | 2005-03-31 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Completely buried, non-ion-paired glutamic acid contributes favorably to the conformational stability of pyrrolidone carboxyl peptidases from hyperthermophiles.
Biochemistry, 45, 2006
|
|
2YZ3
 
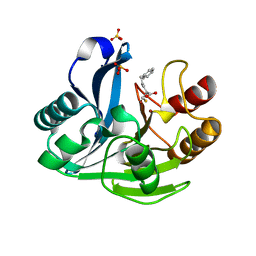 | | Crystallographic Investigation of Inhibition Mode of the VIM-2 Metallo-beta-lactamase from Pseudomonas aeruginosa with Mercaptocarboxylate Inhibitor | | Descriptor: | (S)-2-(MERCAPTOMETHYL)-5-PHENYLPENTANOIC ACID, Metallo-beta-lactamase, SULFATE ION, ... | | Authors: | Yamaguchi, Y, Yamagata, Y, Arakawa, Y, Kurosaki, H. | | Deposit date: | 2007-05-02 | | Release date: | 2008-03-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic investigation of the inhibition mode of a VIM-2 metallo-beta-lactamase from Pseudomonas aeruginosa by a mercaptocarboxylate inhibitor.
J.Med.Chem., 50, 2007
|
|
