5KC5
 
 | | Crystal structure of the Cbln1 C1q domain trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cerebellin-1 | | Authors: | Elegheert, J, Clay, J.E, Siebold, C, Aricescu, A.R. | | Deposit date: | 2016-06-05 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Structural basis for integration of GluD receptors within synaptic organizer complexes.
Science, 353, 2016
|
|
5KC9
 
 | | Crystal structure of the amino-terminal domain (ATD) of iGluR Delta-1 (GluD1) | | Descriptor: | 1,2-ETHANEDIOL, 1,4-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Elegheert, J, Clay, J.E, Siebold, C, Aricescu, A.R. | | Deposit date: | 2016-06-05 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for integration of GluD receptors within synaptic organizer complexes.
Science, 353, 2016
|
|
5KC6
 
 | |
5KC7
 
 | |
7P4G
 
 | |
1NW2
 
 | | The crystal structure of the mutant R82E of Thioredoxin from Alicyclobacillus acidocaldarius | | Descriptor: | ACETATE ION, CACODYLATE ION, THIOREDOXIN, ... | | Authors: | Bartolucci, S, De Simone, G, Galdiero, S, Improta, R, Menchise, V, Pedone, C, Pedone, E, Saviano, M. | | Deposit date: | 2003-02-05 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An integrated structural and computational study of the thermostability of two thioredoxin mutants from Alicyclobacillus acidocaldarius
J.Bacteriol., 185, 2003
|
|
1NSW
 
 | | The Crystal Structure of the K18G Mutant of the thioredoxin from Alicyclobacillus acidocaldarius | | Descriptor: | THIOREDOXIN | | Authors: | Bartolucci, S, De Simone, G, Galdiero, S, Improta, R, Menchise, V, Pedone, C, Pedone, E, Saviano, M. | | Deposit date: | 2003-01-28 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An integrated structural and computational study of the thermostability of two thioredoxin mutants from Alicyclobacillus acidocaldarius
J.Bacteriol., 185, 2003
|
|
3QR6
 
 | |
3SWL
 
 | |
4IS1
 
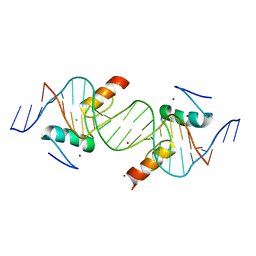 | | Crystal structure of ZNF217 bound to DNA | | Descriptor: | 5'-D(*AP*AP*TP*GP*CP*AP*GP*AP*AP*TP*CP*GP*AP*TP*TP*CP*TP*GP*CP*A)-3', 5'-D(*TP*TP*TP*GP*CP*AP*GP*AP*AP*TP*CP*GP*AP*TP*TP*CP*TP*GP*CP*A)-3', CHLORIDE ION, ... | | Authors: | Vandevenne, M.S, Jacques, D.A, Guss, J.M, Mackay, J.P. | | Deposit date: | 2013-01-16 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New insights into DNA recognition by zinc fingers revealed by structural analysis of the oncoprotein ZNF217.
J.Biol.Chem., 288, 2013
|
|
2I3R
 
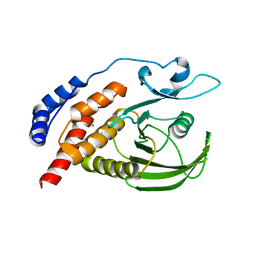 | | Engineered catalytic domain of protein tyrosine phosphatase HPTPbeta | | Descriptor: | CHLORIDE ION, Receptor-type tyrosine-protein phosphatase beta | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M. | | Deposit date: | 2006-08-20 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Engineering the catalytic domain of human protein tyrosine phosphatase beta for structure-based drug discovery.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2I3U
 
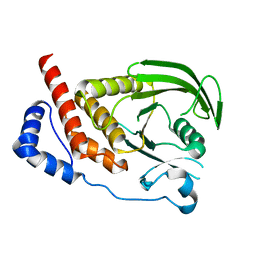 | | Structural studies of protein tyrosine phosphatase beta catalytic domain in complex with inhibitors | | Descriptor: | Receptor-type tyrosine-protein phosphatase beta | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M. | | Deposit date: | 2006-08-20 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Engineering the catalytic domain of human protein tyrosine phosphatase beta for structure-based drug discovery.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2I4E
 
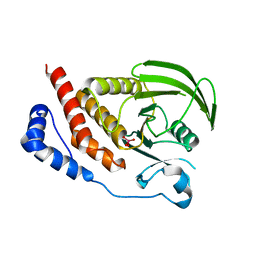 | | Structural studies of protein tyrosine phosphatase beta catalytic domain in complex with inhibitors | | Descriptor: | Receptor-type tyrosine-protein phosphatase beta, VANADATE ION | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M. | | Deposit date: | 2006-08-21 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Engineering the catalytic domain of human protein tyrosine phosphatase beta for structure-based drug discovery.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2I4G
 
 | | Structural studies of protein tyrosine phosphatase beta catalytic domain in complex with a sulfamic acid (soaking experiment) | | Descriptor: | CHLORIDE ION, N-(TERT-BUTOXYCARBONYL)-L-TYROSYL-N-METHYL-4-(SULFOAMINO)-L-PHENYLALANINAMIDE, Receptor-type tyrosine-protein phosphatase beta | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M. | | Deposit date: | 2006-08-21 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Engineering the catalytic domain of human protein tyrosine phosphatase beta for structure-based drug discovery.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2I5X
 
 | | Engineering the PTPbeta catalytic domain with improved crystallization properties | | Descriptor: | (4-ETHYLPHENYL)SULFAMIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M, Klopfenstein, S. | | Deposit date: | 2006-08-26 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Engineering the catalytic domain of human protein tyrosine phosphatase beta for structure-based drug discovery.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
5IRE
 
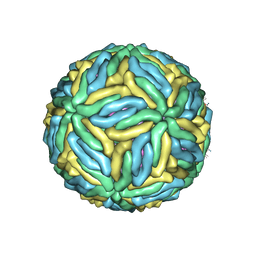 | | The cryo-EM structure of Zika Virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, E protein, M protein | | Authors: | Sirohi, D, Chen, Z, Sun, L, Klose, T, Pierson, T, Rossmann, M, Kuhn, R. | | Deposit date: | 2016-03-13 | | Release date: | 2016-03-30 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The 3.8 angstrom resolution cryo-EM structure of Zika virus.
Science, 352, 2016
|
|
5QUE
 
 | |
5QUH
 
 | |
5QUF
 
 | |
5QUD
 
 | |
5QUB
 
 | |
5QUG
 
 | |
5QUO
 
 | |
5QUC
 
 | |
5QUI
 
 | |
