8EKX
 
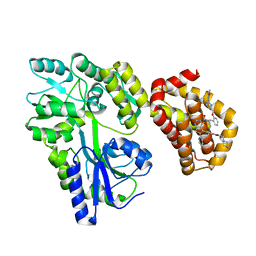 | | Structure of MBP-Mcl-1 in complex with MIK665 | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-4-[2-(4-methylpiperazin-1-yl)ethoxy]phenyl]-6-(4-fluorophenyl)thieno[2,3-d]pyrimidin-4-yl]oxy-3-[2-[[2-(2-methoxyphenyl)pyrimidin-4-yl]methoxy]phenyl]propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Judge, R.A, Judd, A.S, Souers, A.J. | | Deposit date: | 2022-09-22 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Selective MCL-1 inhibitor ABBV-467 is efficacious in tumor models but is associated with cardiac troponin increases in patients.
Commun Med (Lond), 3, 2023
|
|
5UUO
 
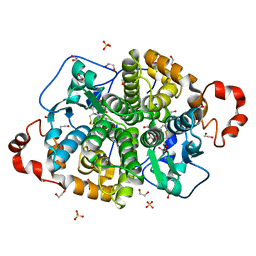 | | Crystal structure of SARO_2595 from Novosphingobium aromaticivorans | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, Glutathione S-transferase-like protein, ... | | Authors: | Bingman, C.A, Kontur, W.S, Olmsted, C.N, Fox, B.G, Donohue, T.J. | | Deposit date: | 2017-02-17 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Novosphingobium aromaticivoransuses a Nu-class glutathioneS-transferase as a glutathione lyase in breaking the beta-aryl ether bond of lignin.
J. Biol. Chem., 293, 2018
|
|
3QUE
 
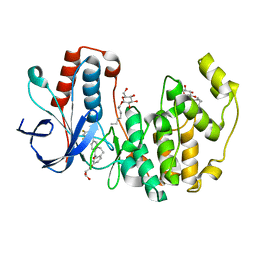 | | Human p38 MAP Kinase in Complex with Skepinone-L | | Descriptor: | 2-[(2,4-difluorophenyl)amino]-7-{[(2R)-2,3-dihydroxypropyl]oxy}-10,11-dihydro-5H-dibenzo[a,d][7]annulen-5-one, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Mayer-Wrangowski, S, Richters, A, Rauh, D. | | Deposit date: | 2011-02-23 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Skepinone-L is a selective p38 mitogen-activated protein kinase inhibitor.
Nat.Chem.Biol., 8, 2012
|
|
5UUN
 
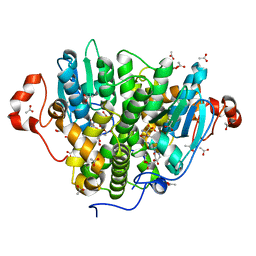 | | Crystal structure of SARO_2595 from Novosphingobium aromaticivorans | | Descriptor: | ACETATE ION, GLUTATHIONE, Glutathione S-transferase-like protein | | Authors: | Bingman, C.A, Kontur, W.S, Olmsted, C.N, Fox, B.G, Donohue, T.J. | | Deposit date: | 2017-02-17 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Novosphingobium aromaticivoransuses a Nu-class glutathioneS-transferase as a glutathione lyase in breaking the beta-aryl ether bond of lignin.
J. Biol. Chem., 293, 2018
|
|
8Q8J
 
 | | Crystal structure of human GPX4-R152H | | Descriptor: | Glutathione peroxidase | | Authors: | Napolitano, V, Mourao, A, Kolonko, M, Bostock, M.J, Sattler, M, Conrad, M, Popowicz, G. | | Deposit date: | 2023-08-18 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human GPX4-R152H
To Be Published
|
|
8Q8N
 
 | | Crystal structure of human GPX4-U46C-I129S-L130S | | Descriptor: | Phospholipid hydroperoxide glutathione peroxidase | | Authors: | Bostock, M.J, Mourao, A, Napolitano, V, Sattler, M, Conrad, M, Popowicz, G. | | Deposit date: | 2023-08-18 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An ultra-rare variant of GPX4 reveals the structural basis to avert neurodegeneration
To Be Published
|
|
5N47
 
 | |
5N48
 
 | |
6R2J
 
 | | Crystal Structure of Pseudomonas stutzeri endoglucanase Cel5A in complex with cellobiose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endoglucanase(Endo-1,4-beta-glucanase)protein, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Dutoit, R, Delsaute, M, Berlemont, R, Van Elder, D, Galleni, M, Bauvois, C. | | Deposit date: | 2019-03-18 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal structure determination of Pseudomonas stutzeri A1501 endoglucanase Cel5A: the search for a molecular basis for glycosynthesis in GH5_5 enzymes.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5I35
 
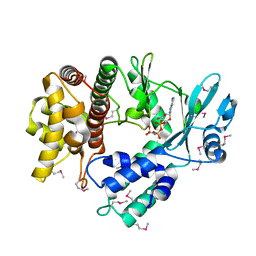 | | Structure of the Human mitochondrial kinase COQ8A R611K with AMPPNP (Cerebellar Ataxia and Ubiquinone Deficiency Through Loss of Unorthodox Kinase Activity) | | Descriptor: | Atypical kinase ADCK3, mitochondrial, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Bingman, C.A, Stefely, J.A, Pagliarini, D.J, Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2016-02-09 | | Release date: | 2016-08-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cerebellar Ataxia and Coenzyme Q Deficiency through Loss of Unorthodox Kinase Activity.
Mol.Cell, 63, 2016
|
|
5D10
 
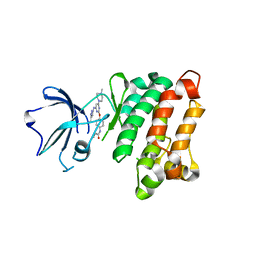 | | Kinase domain of cSrc in complex with RL236 | | Descriptor: | N-[4-({4-(4-methylpiperazin-1-yl)-6-[(5-methyl-1H-pyrazol-3-yl)amino]pyrimidin-2-yl}oxy)phenyl]prop-2-enamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Becker, C, Mayer-Wrangowski, S.C, Julian, E, Rauh, D. | | Deposit date: | 2015-08-03 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Targeting Drug Resistance in EGFR with Covalent Inhibitors: A Structure-Based Design Approach.
J.Med.Chem., 58, 2015
|
|
5D11
 
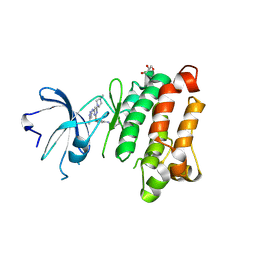 | | Kinase domain of cSrc in complex with RL235 | | Descriptor: | GLYCEROL, N-[3-({4-(4-methylpiperazin-1-yl)-6-[(5-methyl-1H-pyrazol-3-yl)amino]pyrimidin-2-yl}oxy)phenyl]prop-2-enamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Becker, C, Gruetter, C, Engel, J, Rauh, D. | | Deposit date: | 2015-08-03 | | Release date: | 2015-09-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Targeting Drug Resistance in EGFR with Covalent Inhibitors: A Structure-Based Design Approach.
J.Med.Chem., 58, 2015
|
|
3H0E
 
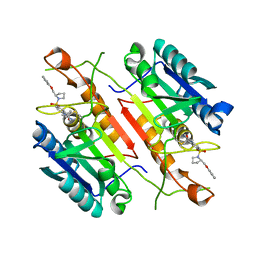 | | 3,4-Dihydropyrimido(1,2-a)indol-10(2H)-ones as Potent Non-Peptidic Inhibitors of Caspase-3 | | Descriptor: | (10S)-3,3-dimethyl-8-{[(2S)-2-(phenoxymethyl)pyrrolidin-1-yl]sulfonyl}-2,3,4,10-tetrahydropyrimido[1,2-a]indol-10-ol, Caspase-3 | | Authors: | Xu, W. | | Deposit date: | 2009-04-09 | | Release date: | 2009-11-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | 3,4-Dihydropyrimido(1,2-a)indol-10(2H)-ones as potent non-peptidic inhibitors of caspase-3
Bioorg.Med.Chem., 17, 2009
|
|
5MGQ
 
 | |
7AKR
 
 | |
7AKS
 
 | |
7AZT
 
 | | X-ray crystallographic structure of (6-4)photolyase from Drosophila melanogaster at room temperature | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, RE11660p | | Authors: | Cellini, A, Wahlgren, W.Y, Henry, L, Westenhoff, S, Pandey, S. | | Deposit date: | 2020-11-17 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | The three-dimensional structure of Drosophila melanogaster (6-4) photolyase at room temperature.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7AYV
 
 | | X-ray crystallographic structure of (6-4)photolyase from Drosophila melanogaster at cryogenic temperature | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, RE11660p, ... | | Authors: | Cellini, A, Wahlgren, W.Y, Henry, L, Westenhoff, S. | | Deposit date: | 2020-11-13 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The three-dimensional structure of Drosophila melanogaster (6-4) photolyase at room temperature.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7AQM
 
 | | ADP-ribosylserine hydrolase ARH3 of Latimeria chalumnae in complex with alpha-1''-O-methyl-ADP-ribose (meADPr) | | Descriptor: | ADP-ribosylhydrolase like 2, Adenosine 5'-diphosphoric acid beta-[(3beta,4beta-dihydroxy-5beta-methoxytetrahydrofuran-2alpha-yl)methyl] estere, MAGNESIUM ION | | Authors: | Rack, J.G.M, Zorzini, V, Ahel, I. | | Deposit date: | 2020-10-22 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic insights into the three steps of poly(ADP-ribosylation) reversal.
Nat Commun, 12, 2021
|
|
7ARW
 
 | | Structure of human ARH3 E41A bound to alpha-NAD+ and magnesium | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADP-ribose glycohydrolase ARH3, ... | | Authors: | Rack, J.G.M, Zorzini, V, Ahel, I. | | Deposit date: | 2020-10-26 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Mechanistic insights into the three steps of poly(ADP-ribosylation) reversal.
Nat Commun, 12, 2021
|
|
1PBW
 
 | | STRUCTURE OF BCR-HOMOLOGY (BH) DOMAIN | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE | | Authors: | Musacchio, A, Cantley, L.C, Harrison, S.C. | | Deposit date: | 1996-10-17 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the breakpoint cluster region-homology domain from phosphoinositide 3-kinase p85 alpha subunit.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
8G08
 
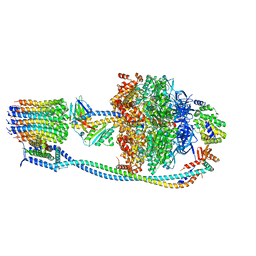 | |
8G09
 
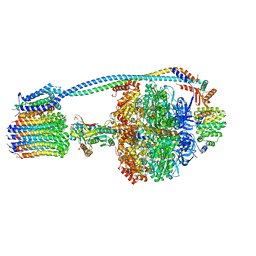 | |
8G0B
 
 | | Cryo-EM structure of TBAJ-876-bound Mycobacterium smegmatis ATP synthase FO region | | Descriptor: | (1R,2S)-1-(6-bromo-2-methoxyquinolin-3-yl)-2-(2,6-dimethoxypyridin-4-yl)-4-(dimethylamino)-1-(2,3,6-trimethoxypyridin-4-yl)butan-2-ol, ATP synthase subunit a, ATP synthase subunit b, ... | | Authors: | Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2023-01-31 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of mycobacterial ATP synthase inhibition by squaramides and second generation diarylquinolines.
Embo J., 42, 2023
|
|
8G0E
 
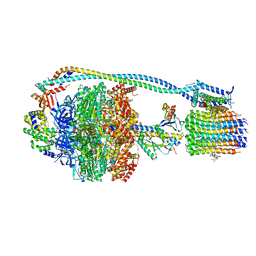 | | Cryo-EM structure of TBAJ-876-bound Mycobacterium smegmatis ATP synthase rotational state 3 | | Descriptor: | (1R,2S)-1-(6-bromo-2-methoxyquinolin-3-yl)-2-(2,6-dimethoxypyridin-4-yl)-4-(dimethylamino)-1-(2,3,6-trimethoxypyridin-4-yl)butan-2-ol, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2023-01-31 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Mechanism of mycobacterial ATP synthase inhibition by squaramides and second generation diarylquinolines.
Embo J., 42, 2023
|
|
